Ann Lab Med.
2023 Nov;43(6):614-619. 10.3343/alm.2023.43.6.614.
Molecular and Clinical Features of Fluconazole Non-susceptible Candida albicans Bloodstream Isolates Recovered in Korean Multicenter Surveillance Studies
- Affiliations
-
- 1Microbiological Analysis Team, Biometrology Group, Korea Research Institute of Standards and Science (KRISS), Daejeon, Korea
- 2Department of Laboratory Medicine, Chonnam National University Medical School, Gwangju, Korea
- 3Department of Laboratory Medicine, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea
- 4Department of Laboratory Medicine, Ajou University School of Medicine, Suwon, Korea
- 5Department of Laboratory Medicine, Jeonbuk National University Medical School and Hospital, Jeonju, Korea
- 6Research Institute of Clinical Medicine of Jeonbuk National University, Biomedical Research Institute of Jeonbuk National University Hospital, Jeonju, Korea
- 7Department of Laboratory Medicine and Research Institute of Bacterial Resistance, Yonsei University College of Medicine, Seoul, Korea
- 8Department of Laboratory Medicine, Pusan National University School of Medicine, Yangsan, Korea
- KMID: 2552039
- DOI: http://doi.org/10.3343/alm.2023.43.6.614
Abstract
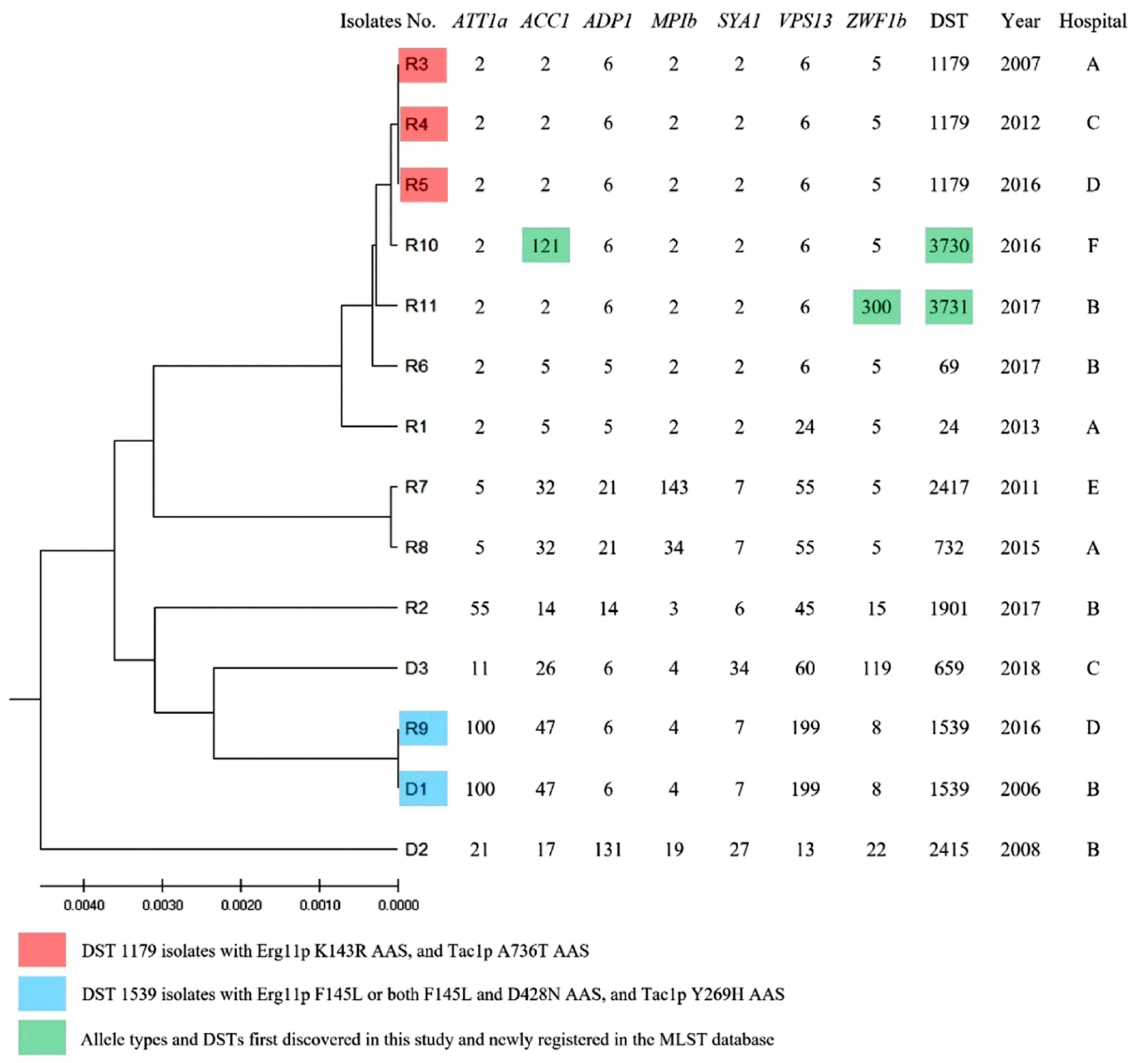
- Acquired fluconazole resistance (FR) in bloodstream infection (BSI) isolates of Candida albicans is rare. We investigated the FR mechanisms and clinical features of 14 fluconazole non-susceptible (FNS; FR and fluconazole-susceptible dose-dependent) BSI isolates of C. albicans recovered from Korean multicenter surveillance studies during 2006–2021. Mutations causing amino acid substitutions (AASs) in the drug-target gene ERG11 and the FR-associated transcription factor genes TAC1 , MRR1, and UPC2 of the 14 FNS isolates were compared with those of 12 fluconazole-susceptible isolates. Of the 14 FNS isolates, eight and seven had Erg11p (K143R, F145L, or G464S) and Tac1p (T225A, R673L, A736T, or A736V) AASs, respectively, which were previously described in FR isolates. Novel Erg11p, Tac1p, and Mrr1p AASs were observed in two, four, and one FNS isolates, respectively. Combined Erg11p and Tac1p AASs were observed in seven FNS isolates. None of the FR-associated Upc2p AASs were detected. Of the 14 patients, only one had previous azole exposure, and the 30-day mortality rate was 57.1% (8/14). Our data show that Erg11p and Tac1p AASs are likely to contribute to FR in C. albicans BSI isolates in Korea and that most FNS C. albicans BSIs develop without azole exposure.
Keyword
Figure
Reference
-
1. Pfaller MA, Diekema DJ, Gibbs DL, Newell VA, Ellis D, Tullio V, et al. Results from the ARTEMIS DISK Global Antifungal Surveillance Study, 1997 to 2007: a 10.5-year analysis of susceptibilities of Candida species to fluconazole and voriconazole as determined by CLSI standardized disk diffusion. J Clin Microbiol. 2010; 48:1366–77. DOI: 10.1128/JCM.02117-09. PMID: 20164282. PMCID: PMC2849609.
Article2. Nishimoto AT, Sharma C, Rogers PD. 2020; Molecular and genetic basis of azole antifungal resistance in the opportunistic pathogenic fungus Candida albicans. J Antimicrob Chemother. 75:257–70. DOI: 10.1093/jac/dkz400. PMID: 31603213. PMCID: PMC8204710.3. Collins LM, Moore R, Sobel JD. 2020; Prognosis and long-term outcome of women with idiopathic recurrent vulvovaginal candidiasis caused by albicans. J Low Genit Tract Dis. 24:48–52. DOI: 10.1097/LGT.0000000000000496. PMID: 31860575.
Article4. Pfaller MA, Andes D, Diekema DJ, Espinel-Ingroff A, Sheehan D. CLSI Subcommittee for Antifungal Susceptibility Testing. 2010; Wild-type MIC distributions, epidemiological cutoff values and species-specific clinical breakpoints for fluconazole and Candida: time for harmonization of CLSI and EUCAST broth microdilution methods. Drug Resist Updat. 13:180–95. DOI: 10.1016/j.drup.2010.09.002. PMID: 21050800.5. Perlin DS, Wiederhold NP. 2017; Culture-independent molecular methods for detection of antifungal resistance mechanisms and fungal identification. J Infect Dis. 216(S3):S458–65. DOI: 10.1093/infdis/jix121. PMID: 28911041.6. Morio F, Pagniez F, Besse M, Gay-Andrieu F, Miegeville M, Le Pape P. 2013; Deciphering azole resistance mechanisms with a focus on transcription factor-encoding genes TAC1, MRR1 and UPC2 in a set of fluconazole-resistant clinical isolates of Candida albicans. Int J Antimicrob Agents. 42:410–5. DOI: 10.1016/j.ijantimicag.2013.07.013. PMID: 24051054.7. Dunkel N, Blass J, Rogers PD, Morschhäuser J. 2008; Mutations in the multi-drug resistance regulator MRR1, followed by loss of heterozygosity, are the main cause of MDR1 overexpression in fluconazole-resistant Candida albicans strains. Mol Microbiol. 69:827–40. DOI: 10.1111/j.1365-2958.2008.06309.x. PMID: 18577180. PMCID: PMC2678921.8. Pfaller MA, Diekema DJ, Sheehan DJ. 2006; Interpretive breakpoints for fluconazole and Candida revisited: a blueprint for the future of antifungal susceptibility testing. Clin Microbiol Rev. 19:435–47. DOI: 10.1128/CMR.19.2.435-447.2006. PMID: 16614256. PMCID: PMC1471993.9. Lockhart SR, Iqbal N, Cleveland AA, Farley MM, Harrison LH, Bolden CB, et al. 2012; Species identification and antifungal susceptibility testing of Candida bloodstream isolates from population-based surveillance studies in two U.S. cities from 2008 to 2011. J Clin Microbiol. 50:3435–42. DOI: 10.1128/JCM.01283-12. PMID: 22875889. PMCID: PMC3486211.10. Castanheira M, Deshpande LM, Davis AP, Rhomberg PR, Pfaller MA. 2017; Monitoring antifungal resistance in a global collection of invasive yeasts and molds: application of CLSI epidemiological cutoff values and whole-genome sequencing analysis for detection of azole resistance in Candida albicans. Antimicrob Agents Chemother. 61:e00906–17. DOI: 10.1128/AAC.00906-17. PMID: 28784671. PMCID: PMC5610521.11. Lee HS, Shin JH, Choi MJ, Won EJ, Kee SJ, Kim SH, et al. 2017; Comparison of the Bruker Biotyper and VITEK MS matrix-assisted laser desorption/ionization time-of-flight mass spectrometry systems using a formic acid extraction method to identify common and uncommon yeast isolates. Ann Lab Med. 37:223–30. DOI: 10.3343/alm.2017.37.3.223. PMID: 28224768. PMCID: PMC5339094.
Article12. Jeon S, Shin JH, Lim HJ, Choi MJ, Byun SA, Lee D, et al. 2021; Disk diffusion susceptibility testing for the rapid detection of fluconazole resistance in Candida isolates. Ann Lab Med. 41:559–67. DOI: 10.3343/alm.2021.41.6.559. PMID: 34108283. PMCID: PMC8203430.
Article13. Park S, Perlin DS. 2005; Establishing surrogate markers for fluconazole resistance in Candida albicans. Microb Drug Resist. 11:232–8. DOI: 10.1089/mdr.2005.11.232. PMID: 16201925.14. Shin JH, Bougnoux ME, d'Enfert C, Kim SH, Moon CJ, Joo MY, et al. 2011; Genetic diversity among Korean Candida albicans bloodstream isolates: assessment by multilocus sequence typing and restriction endonuclease analysis of genomic DNA by use of BssHII. J Clin Microbiol. 49:2572–7. DOI: 10.1128/JCM.02153-10. PMID: 21562112. PMCID: PMC3147862.
Article15. Kwon YJ, Won EJ, Jeong SH, Shin KS, Shin JH, Kim YR, et al. 2021; Dynamics and predictors of mortality due to candidemia caused by different Candida species: comparison of intensive care unit-associated candidemia (ICUAC) and non-ICUAC. J Fungi (Basel). 7:597. DOI: 10.3390/jof7080597. PMID: 34436136. PMCID: PMC8397010.
Article16. Flowers SA, Colón B, Whaley SG, Schuler MA, Rogers PD. 2015; Contribution of clinically derived mutations in ERG11 to azole resistance in Candida albicans. Antimicrob Agents Chemother. 59:450–60. DOI: 10.1128/AAC.03470-14. PMID: 25385095. PMCID: PMC4291385.
Article17. Xu Y, Chen L, Li C. 2008; Susceptibility of clinical isolates of Candida species to fluconazole and detection of Candida albicans ERG11 mutations. J Antimicrob Chemother. 61:798–804. DOI: 10.1093/jac/dkn015. PMID: 18218640.
Article18. Coste A, Selmecki A, Forche A, Diogo D, Bougnoux ME, d'Enfert C, et al. 2007; Genotypic evolution of azole resistance mechanisms in sequential Candida albicans isolates. Eukaryot Cell. 6:1889–904. DOI: 10.1128/EC.00151-07. PMID: 17693596. PMCID: PMC2043391.
Article19. Coste AT, Crittin J, Bauser C, Rohde B, Sanglard D. 2009; Functional analysis of cis- and trans-acting elements of the Candida albicans CDR2 promoter with a novel promoter reporter system. Eukaryot Cell. 8:1250–67. DOI: 10.1128/EC.00069-09. PMID: 19561319. PMCID: PMC2725566.
Article20. Jensen RH, Astvad KM, Silva LV, Sanglard D, Jørgensen R, Nielsen KF, et al. 2015; Stepwise emergence of azole, echinocandin and amphotericin B multidrug resistance in vivo in Candida albicans orchestrated by multiple genetic alterations. J Antimicrob Chemother. 70:2551–5. DOI: 10.1093/jac/dkv140. PMID: 26017038. PMCID: PMC4553713.21. Flowers SA, Barker KS, Berkow EL, Toner G, Chadwick SG, Gygax SE, et al. 2012; Gain-of-function mutations in UPC2 are a frequent cause of ERG11 upregulation in azole-resistant clinical isolates of Candida albicans. Eukaryot Cell. 11:1289–99. DOI: 10.1128/EC.00215-12. PMID: 22923048. PMCID: PMC3485914.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Species Distribution and Susceptibility to Azole Antifungals of Candida Bloodstream Isolates from Eight University Hospitals in Korea
- Antifungal Susceptibilities to Fulconazole and Itraconazole for Candida Species Recovered from Blood Cultures over a 5-Year Period
- Efficacy of Fluconazole in The Treatment of Candida albicans Keratitis in Rabbits
- Candida Species Isolated from Clinical Specimens and Medical Personnel
- Antifungal Susceptibility Testing with Etest for Candida Species Isolated from Patients with Oral Candidiasis