Ann Lab Med.
2023 Sep;43(5):434-442. 10.3343/alm.2023.43.5.434.
Comparison of Nasal Swabs, Nasopharyngeal Swabs, and Saliva Samples for the Detection of SARS-CoV-2 and other Respiratory Virus Infections
- Affiliations
-
- 1Division of Infectious Diseases, Department of Internal Medicine, Hallym University Dongtan Sacred Heart Hospital, Hallym University College of Medicine, Hwaseong, Korea
- 2Departments of Laboratory Medicine, Hallym University Dongtan Sacred Heart Hospital, Hallym University College of Medicine, Hwaseong, Korea
- 3Departments Pediatrics, Hallym University Dongtan Sacred Heart Hospital, Hallym University College of Medicine, Hwaseong, Korea
- 4Department of Laboratory Medicine, Kangdong Sacred Heart Hospital, Seoul, Korea
- KMID: 2551991
- DOI: http://doi.org/10.3343/alm.2023.43.5.434
Abstract
- Background
Nasal swabs and saliva samples are being considered alternatives to nasopharyngeal swabs (NPSs) for detecting severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2); however, few studies have compared the usefulness of nasal swabs, NPSs, and saliva samples for detecting SARS-CoV-2 and other respiratory virus infections. We compared the positivity rates and concentrations of viruses detected in nasal swabs, NPSs, and saliva samples using cycle threshold (Ct) values from real-time PCR tests for respiratory viruses.
Methods
In total, 236 samples (48 five-rub and 10 10-rub nasal swabs, 96 NPSs collected using two different products, 48 saliva swabs, and 34 undiluted saliva samples) from 48 patients (34 patients with SARS-CoV-2 and 14 with other respiratory virus infections) and 40 samples from eight healthy controls were obtained. The PCR positivity and Ct values were compared using Allplex Respiratory Panels 1/2/3 and Allplex SARS-CoV-2 real-time PCR.
Results
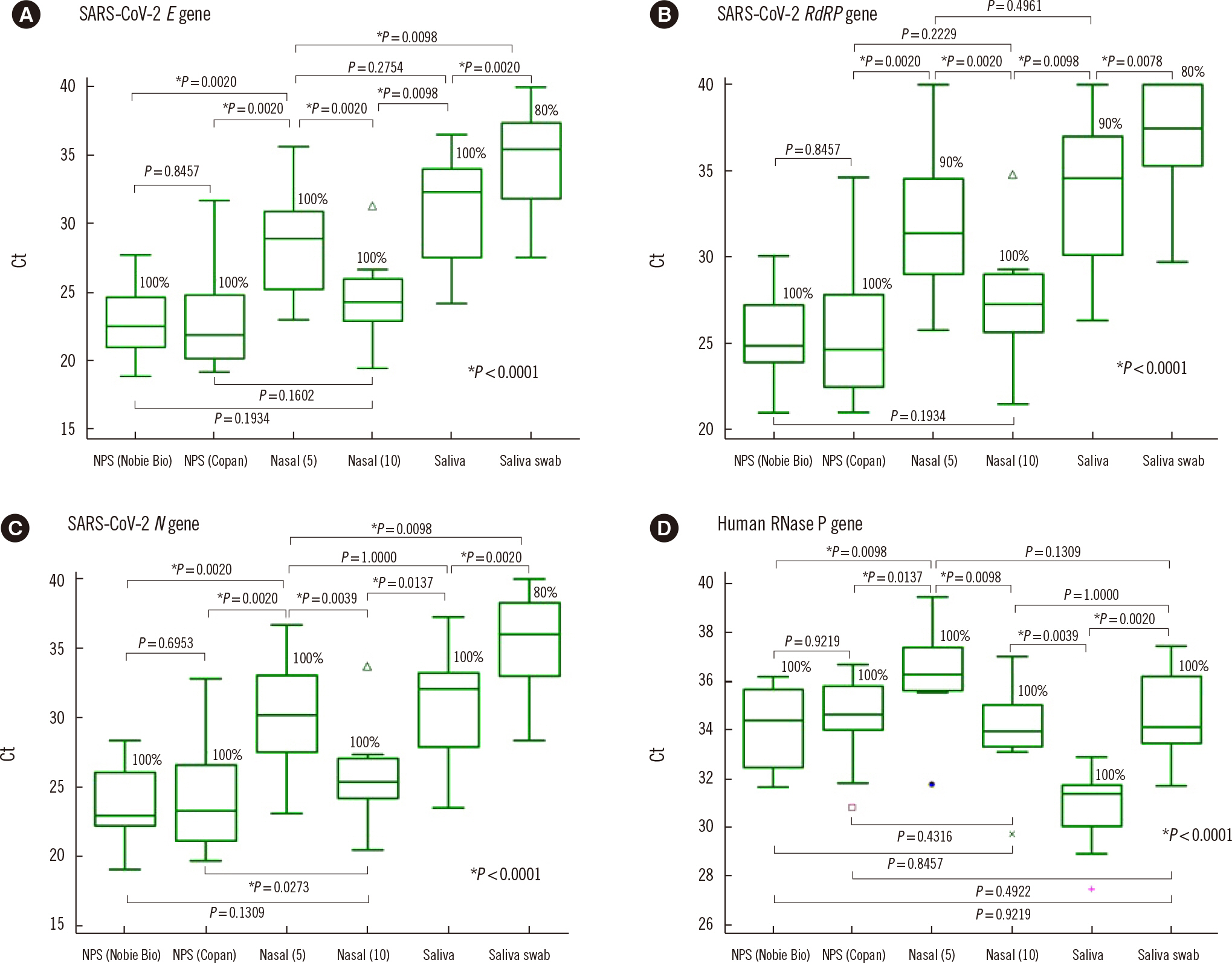
NPSs showed the lowest Ct values (indicating the highest virus concentrations); however, nasal and saliva samples yielded positive results for SARS-CoV-2 and other respiratory viruses. The median Ct value for SARS-CoV-2 E gene PCR using nasal swab samples collected with 10 rubs was significantly different from that obtained using nasal swabs collected with five rubs (Ct=24.3 vs. 28.9; P=0.002), but not from that obtained using NPSs.
Conclusions
Our results confirm that the NPS is the best sample type for detecting respiratory viruses, but nasal swabs and saliva samples can be alternatives to NPSs. Vigorously and sufficiently rubbed nasal swabs can provide SARS-CoV-2 concentrations similar to those obtained with NPSs.
Keyword
Figure
Reference
-
1. Miller JM, Binnicker MJ, Campbell S, Carroll KC, Chapin KC, Gilligan PH, et al. 2018; A guide to utilization of the microbiology laboratory for diagnosis of infectious diseases: 2018 update by the Infectious Diseases Society of America and the American Society for Microbiology. Clin Infect Dis. 67:e1–e94. DOI: 10.1093/cid/ciy381. PMID: 29955859. PMCID: PMC7108105.2. Hong KH, Kim GJ, Roh KH, Sung H, Lee J, Kim SY, et al. 2022; Update of guidelines for laboratory diagnosis of COVID-19 in Korea. Ann Lab Med. 42:391–7. DOI: 10.3343/alm.2022.42.4.391. PMID: 35177559. PMCID: PMC8859556.
Article3. Oh SM, Jeong H, Chang E, Choe PG, Kang CK, Park WB, et al. 2021; Clinical application of the standard Q COVID-19 Ag test for the detection of SARS-CoV-2 infection. J Korean Med Sci. 36:e101. DOI: 10.3346/jkms.2021.36.e101. PMID: 33847084. PMCID: PMC8042480.
Article4. Corman VM, Haage VC, Bleicker T, Schmidt ML, Mühlemann B, Zuchowski M, et al. 2021; Comparison of seven commercial SARS-CoV-2 rapid point-of-care antigen tests: a single-centre laboratory evaluation study. Lancet Microbe. 2:e311–9. DOI: 10.1016/S2666-5247(21)00056-2. PMID: 33846704.
Article5. Torjesen I. 2021; Covid-19: How the UK is using lateral flow tests in the pandemic. BMJ. 372:n287. DOI: 10.1136/bmj.n287. PMID: 33541908.
Article6. Lindner AK, Nikolai O, Kausch F, Wintel M, Hommes F, Gertler M, et al. 2021; Head-to-head comparison of SARS-CoV-2 antigen-detecting rapid test with self-collected nasal swab versus professional-collected nasopharyngeal swab. Eur Respir J. 57:2003961. DOI: 10.1183/13993003.03961-2020. PMID: 33303544. PMCID: PMC7736752.7. Jegerlehner S, Suter-Riniker F, Jent P, Bittel P, Nagler M. 2022; Diagnostic accuracy of SARS-CoV-2 saliva antigen testing in a real-life clinical setting. Int J Infect Dis. 119:38–40. DOI: 10.1016/j.ijid.2022.03.037. PMID: 35364282. PMCID: PMC8964446.
Article8. Alonaizan F, AlHumaid J, AlJindan R, Bedi S, Dardas H, Abdulfattah D, et al. 2022; Sensitivity and specificity of rapid SARS-CoV-2 antigen detection using different sampling methods: a clinical unicentral study. Int J Environ Res Public Health. 19:6836. DOI: 10.3390/ijerph19116836. PMID: 35682419. PMCID: PMC9180118.
Article9. Homza M, Zelena H, Janosek J, Tomaskova H, Jezo E, Kloudova A, et al. 2021; Performance of seven SARS-CoV-2 self-tests based on saliva, anterior nasal and nasopharyngeal swabs corrected for infectiousness in real-life conditions: a cross-sectional test accuracy study. Diagnostics (Basel). 11:1567. DOI: 10.3390/diagnostics11091567. PMID: 34573909. PMCID: PMC8466378.
Article10. Alemany A, Millat-Martinez P, Ouchi D, Corbacho-Monné M, Bordoy AE, Esteban C, et al. 2021; Self-collected mid-nasal swabs and saliva specimens, compared with nasopharyngeal swabs, for SARS-CoV-2 detection in mild COVID-19 patients. J Infect. 83:709–37. DOI: 10.1016/j.jinf.2021.09.012. PMID: 34537322. PMCID: PMC8444446.
Article11. Savela ES, Viloria Winnett A, Romano AE, Porter MK, Shelby N, Akana R, et al. 2022; Quantitative SARS-CoV-2 viral-load curves in paired saliva samples and nasal swabs inform appropriate respiratory sampling site and analytical test sensitivity required for earliest viral detection. J Clin Microbiol. 60:e0178521. DOI: 10.1128/jcm.01785-21. PMID: 34911366. PMCID: PMC8849374.
Article12. Lai J, German J, Hong F, Tai SS, McPhaul KM, Milton DK, et al. 2022; Comparison of saliva and midturbinate swabs for detection of SARS-CoV-2. Microbiol Spectr. 10:e0012822. DOI: 10.1128/spectrum.00128-22. PMID: 35311575. PMCID: PMC9045394.
Article13. McLennan K, Barton E, Lang C, Adams IR, McAllister G, Reijns MAM, et al. 2022; User acceptability of saliva and gargle samples for identifying COVID-19 positive high-risk workers and household contacts. Diagn Microbiol Infect Dis. 104:115732. DOI: 10.1016/j.diagmicrobio.2022.115732. PMID: 35728458. PMCID: PMC9132684.
Article14. Kim YG, Yun SG, Kim MY, Park K, Cho CH, Yoon SY, et al. 2017; Comparison between saliva and nasopharyngeal swab specimens for detection of respiratory viruses by multiplex reverse transcription-PCR. J Clin Microbiol. 55:226–33. DOI: 10.1128/JCM.01704-16. PMID: 27807150. PMCID: PMC5228234.
Article15. Hou N, Wang K, Zhang H, Bai M, Chen H, Song W, et al. 2020; Comparison of detection rate of 16 sampling methods for respiratory viruses: a Bayesian network meta-analysis of clinical data and systematic review. BMJ Glob Health. 5:e003053. DOI: 10.1136/bmjgh-2020-003053. PMID: 33168521. PMCID: PMC7654123.
Article16. Woodall CA, Thornton HV, Anderson EC, Ingle SM, Muir P, Vipond B, et al. 2021; Prospective study of the performance of parent-collected nasal and saliva swab samples, compared with nurse-collected swab samples, for the molecular detection of respiratory microorganisms. Microbiol Spectr. 9:e0016421. DOI: 10.1128/Spectrum.00164-21. PMID: 34756077. PMCID: PMC8579848.
Article17. To KKW, Lu L, Yip CC, Poon RWS, Fung AMY, Cheng A, et al. 2017; Additional molecular testing of saliva specimens improves the detection of respiratory viruses. Emerg Microbes Infect. 6:e49. DOI: 10.1038/emi.2017.35. PMID: 28588283. PMCID: PMC5520312.
Article18. Wang WK, Chen SY, Liu IJ, Chen YC, Chen HL, Yang CF, et al. 2004; Detection of SARS-associated coronavirus in throat wash and saliva in early diagnosis. Emerg Infect Dis. 10:1213–9. DOI: 10.3201/eid1007.031113. PMID: 15324540. PMCID: PMC3323313.
Article19. Bennett S, Davidson RS, Gunson RN. 2017; Comparison of gargle samples and throat swab samples for the detection of respiratory pathogens. J Virol Methods. 248:83–6. DOI: 10.1016/j.jviromet.2017.06.010. PMID: 28633963.
Article20. Abdollahi A, Salarvand S, Ghalehtaki R, Jafarzadeh B, Beigmohammadi MT, Ghiasvand F, et al. 2022; The role of saliva PCR assay in the diagnosis of COVID-19. J Infect Dev Ctries. 16:5–9. DOI: 10.3855/jidc.15239. PMID: 35192515.
Article21. Marais G, Hsiao NY, Iranzadeh A, Doolabh D, Joseph R, Enoch A, et al. 2022; Improved oral detection is a characteristic of Omicron infection and has implications for clinical sampling and tissue tropism. J Clin Virol. 152:105170. DOI: 10.1016/j.jcv.2022.105170. PMID: 35525108.
Article22. Berenger BM, Fonseca K, Schneider AR, Hu J, Zelyas N. 2022; Clinical evaluation of nasopharyngeal, midturbinate nasal and oropharyngeal swabs for the detection of SARS-CoV-2. Diagn Microbiol Infect Dis. 102:115618. DOI: 10.1016/j.diagmicrobio.2021.115618. PMID: 35007959. PMCID: PMC8675123.
Article23. Iwasaki S, Fujisawa S, Nakakubo S, Kamada K, Yamashita Y, Fukumoto T, et al. 2020; Comparison of SARS-CoV-2 detection in nasopharyngeal swab and saliva. J Infect. 81:e145–7. DOI: 10.1016/j.jinf.2020.05.071. PMID: 32504740. PMCID: PMC7270800.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Viral Load Kinetics of SARS-CoV-2Infection in Saliva in Korean Patients:a Prospective Multi-center Comparative Study
- Evaluation of Three Multiplex Realtime Reverse Transcription PCR Assays for Simultaneous Detection of SARS-CoV-2, Influenza A/B, and Respiratory Syncytial Virus in Nasopharyngeal Swabs
- Persistent severe acute respiratory syndrome coronavirus 2 detection after resolution of coronavirus disease 2019-associated symptoms/signs
- Nucleic acid-based molecular diagnostic testing of SARS-CoV-2 using self-collected saliva specimens
- Serial Screening for SARS-CoV-2 in Rectal Swabs of Symptomatic COVID-19 Patients