Ann Lab Med.
2023 Jul;43(4):328-336. 10.3343/alm.2023.43.4.328.
Development of a Next-generation Sequencing-based Gene Panel Test to Detect Measurable Residual Disease in Acute Myeloid Leukemia
- Affiliations
-
- 1Department of Laboratory Medicine, Yonsei University College of Medicine, Severance Hospital, Seoul, Korea
- 2Division of Hematology, Department of Internal Medicine, Yonsei University College of Medicine, Severance Hospital, Seoul, Korea
- 3Department of Laboratory Medicine, Graduate School of Medical Science, Brain Korea 21 PLUS Project, Yonsei University College of Medicine, Seoul, Korea
- 4Dxome Co. Ltd., Seongnam, Korea
- KMID: 2551950
- DOI: http://doi.org/10.3343/alm.2023.43.4.328
Abstract
- Background
AML is a heterogeneous disease, and despite intensive therapy, recurrence is still high in AML patients who achieve the criterion for cytomorphologic remission (residual tumor burden [measurable residual disease, MRD]<5%). This study aimed to develop a targeted next-generation sequencing (NGS) panel to detect MRD in AML patients and validate its performance.
Methods
We designed an error-corrected, targeted MRD-NGS panel without using physical molecular barcodes, including 24 genes. Fifty-four bone marrow and peripheral blood samples from 23 AML patients were sequenced using the panel. The panel design was validated using reference material, and accuracy was assessed using droplet digital PCR.
Results
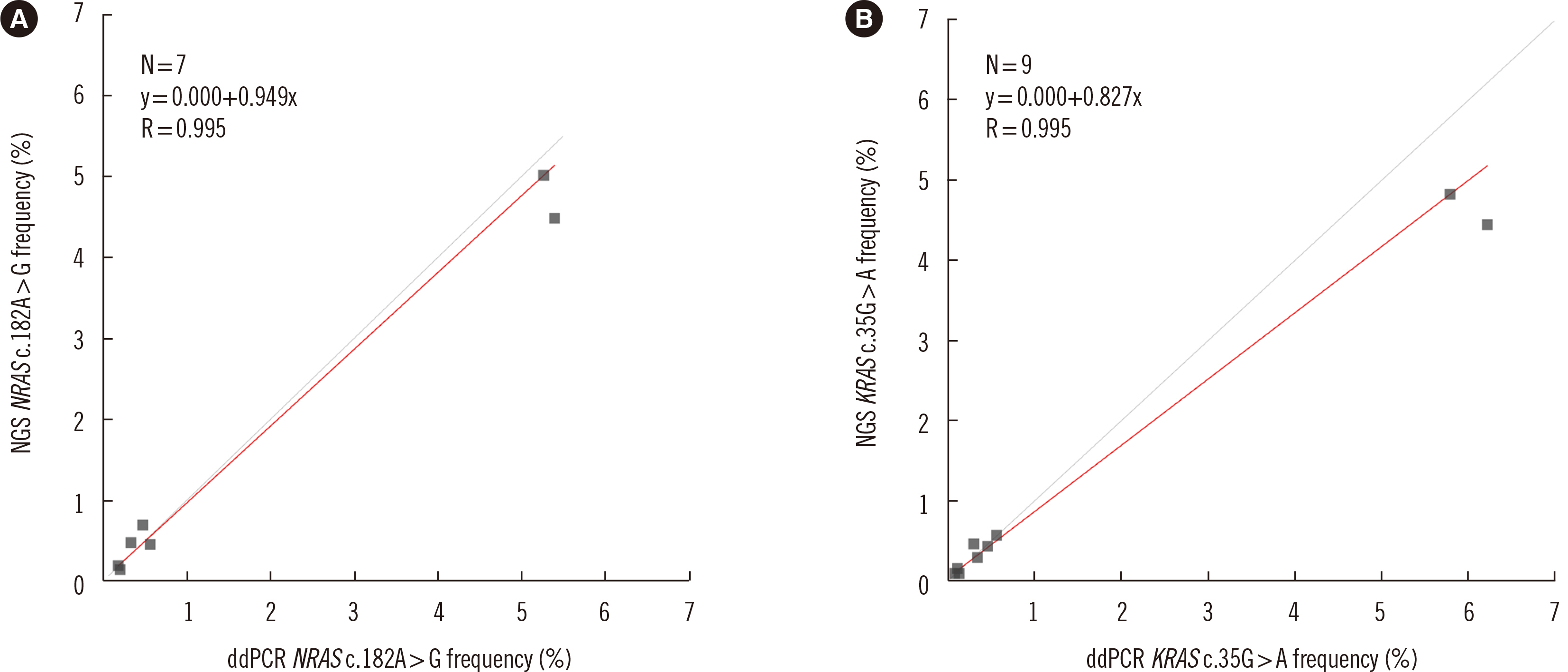
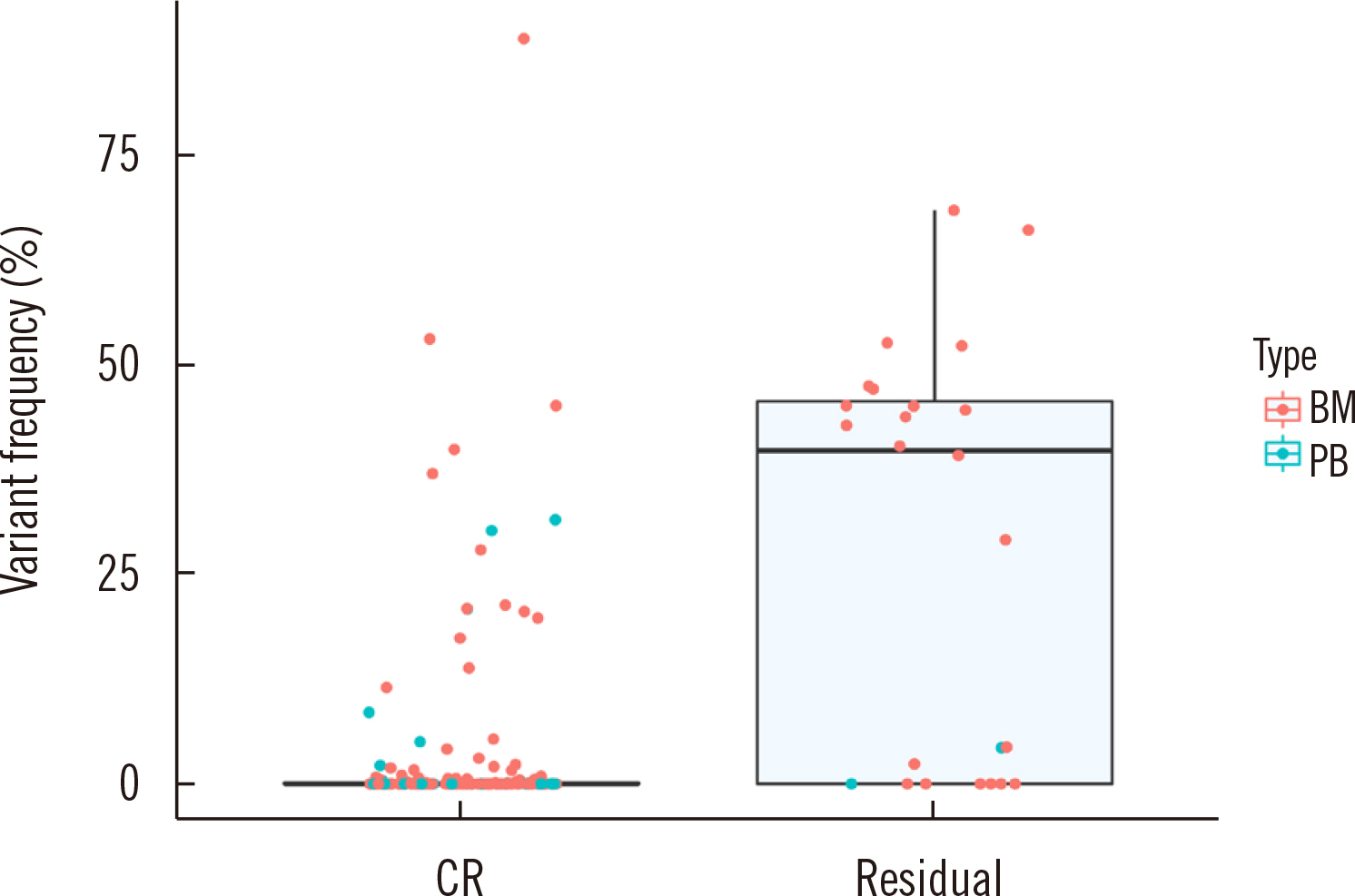
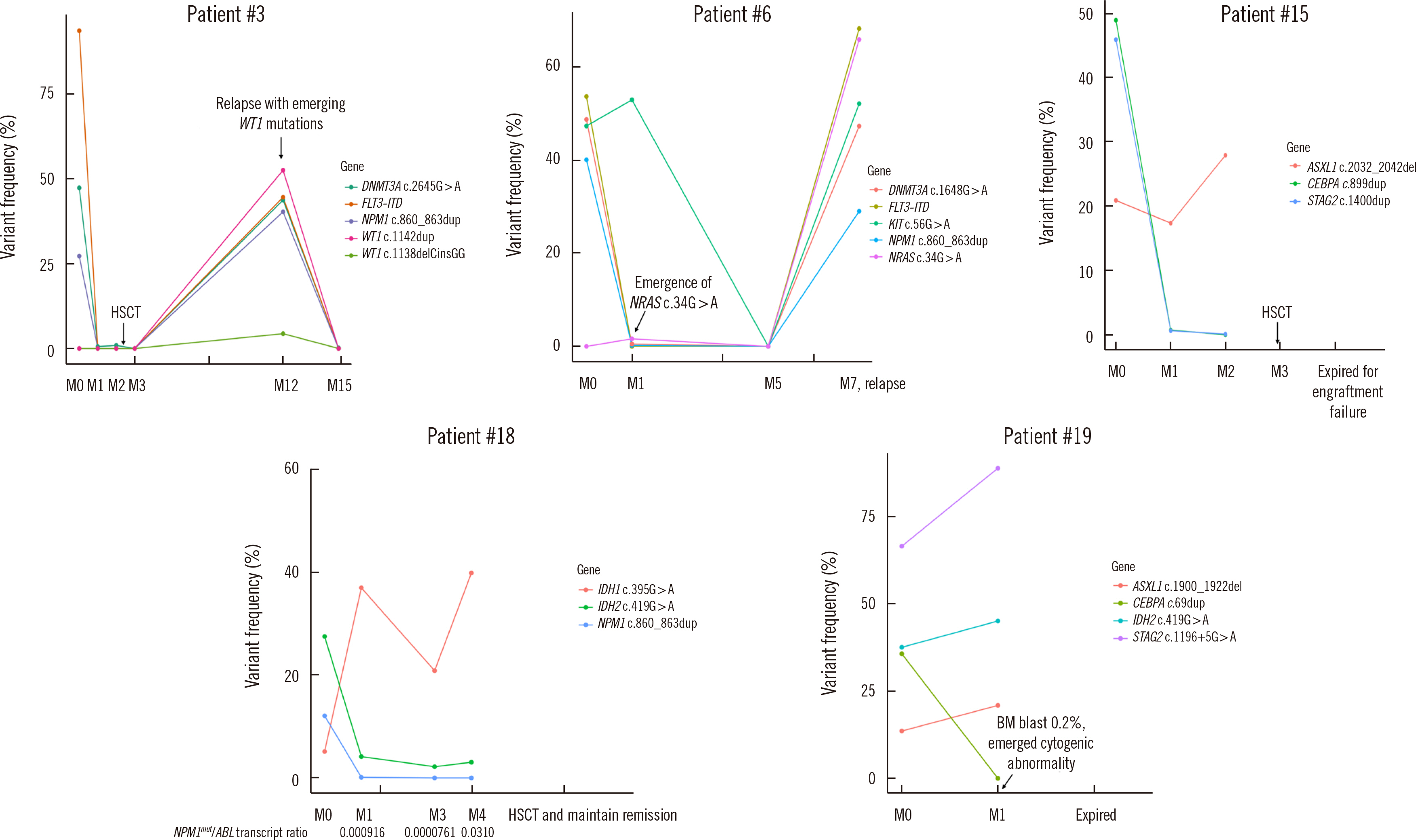
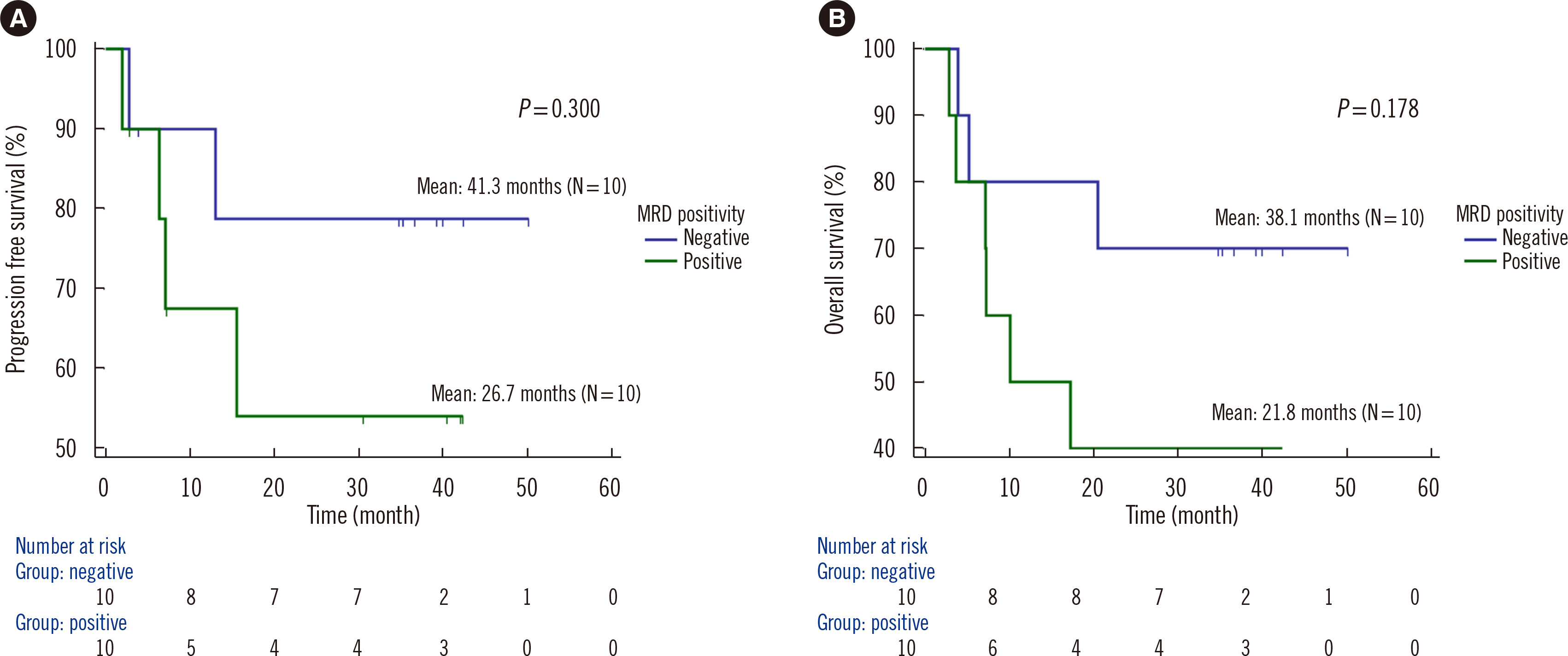
Dilution tests showed excellent linearity and a strong correlation between expected and observed clonal frequencies (R>0.99). The test reproducibly detected MRD in three dilution series samples, with a sensitivity of 0.25% for single-nucleotide variants. More than half of samples from patients with morphologic remission after one month of chemotherapy had detectable mutations. NGS-MRD positivity for samples collected after one month of chemotherapy tended to be associated with poor overall survival and progression-free survival.
Conclusions
Our highly sensitive and accurate NGS-MRD panel can be readily used to monitor most AML patients in clinical practice, including patients without gene rearrangement. In addition, this NGS-MRD panel may allow the detection of newly emerging clones during clinical relapse, leading to more reliable prognoses of AML.
Figure
Cited by 1 articles
-
Measurable Residual Disease Testing Using Next-Generation Sequencing in Acute Myeloid Leukemia
Seon Young Kim, Hee Jin Huh
Ann Lab Med. 2023;43(4):323-324. doi: 10.3343/alm.2023.43.4.323.
Reference
-
1. Morita K, Wang F, Jahn K, Hu T, Tanaka T, Sasaki Y, et al. 2020; Clonal evolution of acute myeloid leukemia revealed by high-throughput single-cell genomics. Nat Commun. 11:5327. DOI: 10.1038/s41467-020-19119-8. PMID: 33087716. PMCID: PMC7577981.
Article2. Tsai CH, Tang JL, Tien FM, Kuo YY, Wu DC, Lin CC, et al. 2021; Clinical implications of sequential MRD monitoring by NGS at 2 time points after chemotherapy in patients with AML. Blood Adv. 5:2456–66. DOI: 10.1182/bloodadvances.2020003738. PMID: 33999144. PMCID: PMC8152512.
Article3. Döhner H, Weisdorf DJ, Bloomfield CD. 2015; Acute myeloid leukemia. N Engl J Med. 373:1136–52. DOI: 10.1056/NEJMra1406184. PMID: 26376137.
Article4. Schuurhuis GJ, Heuser M, Freeman S, Béné MC, Buccisano F, Cloos J, et al. 2018; Minimal/measurable residual disease in AML: a consensus document from the European LeukemiaNet MRD Working Party. Blood. 131:1275–91. DOI: 10.1182/blood-2017-09-801498. PMID: 29330221. PMCID: PMC5865231.
Article5. Panuzzo C, Jovanovski A, Ali MS, Cilloni D, Pergolizzi B. 2022; Revealing the mysteries of acute myeloid leukemia: from quantitative PCR through next-generation sequencing and systemic metabolomic profiling. J Clin Med. 11:483. DOI: 10.3390/jcm11030483. PMID: 35159934. PMCID: PMC8836582.
Article6. Zhong Y, Xu F, Wu J, Schubert J, Li MM. 2021; Application of next generation sequencing in laboratory medicine. Ann Lab Med. 41:25–43. DOI: 10.3343/alm.2021.41.1.25. PMID: 32829577. PMCID: PMC7443516.
Article7. Li S, Garrett-Bakelman FE, Chung SS, Sanders MA, Hricik T, Rapaport F, et al. 2016; Distinct evolution and dynamics of epigenetic and genetic heterogeneity in acute myeloid leukemia. Nat Med. 22:792–9. DOI: 10.1038/nm.4125. PMID: 27322744. PMCID: PMC4938719.
Article8. Krönke J, Bullinger L, Teleanu V, Tschürtz F, Gaidzik VI, Kühn MW, et al. 2013; Clonal evolution in relapsed NPM1-mutated acute myeloid leukemia. Blood. 122:100–8. DOI: 10.1182/blood-2013-01-479188. PMID: 23704090.9. Parkin B, Ouillette P, Li Y, Keller J, Lam C, Roulston D, et al. 2013; Clonal evolution and devolution after chemotherapy in adult acute myelogenous leukemia. Blood. 121:369–77. DOI: 10.1182/blood-2012-04-427039. PMID: 23175688. PMCID: PMC3653567.
Article10. Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T, et al. 2017; Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 129:424–47. DOI: 10.1182/blood-2016-08-733196. PMID: 27895058. PMCID: PMC5291965.
Article11. Chen G, Mosier S, Gocke CD, Lin MT, Eshleman JR. 2014; Cytosine deamination is a major cause of baseline noise in next-generation sequencing. Mol Diagn Ther. 18:587–93. DOI: 10.1007/s40291-014-0115-2. PMID: 25091469. PMCID: PMC4175022.
Article12. Jennings LJ, Arcila ME, Corless C, Kamel-Reid S, Lubin IM, Pfeifer J, et al. 2017; Guidelines for validation of next-generation sequencing-based oncology panels: a joint consensus recommendation of the association for molecular pathology and College of American Pathologists. J Mol Diagn. 19:341–65. DOI: 10.1016/j.jmoldx.2017.01.011. PMID: 28341590. PMCID: PMC6941185.13. Bolger AM, Lohse M, Usadel B. 2014; Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–20. DOI: 10.1093/bioinformatics/btu170. PMID: 24695404. PMCID: PMC4103590.
Article14. Li H. 2014; Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics. 30:2843–51. DOI: 10.1093/bioinformatics/btu356. PMID: 24974202. PMCID: PMC4271055.
Article15. Lee KS, Seo J, Lee CK, Shin S, Choi Z, Min S, et al. 2022; Analytical and clinical validation of cell-free circulating tumor DNA assay for the estimation of tumor mutational burden. Clin Chem. 68:1519–28. DOI: 10.1093/clinchem/hvac146. PMID: 36306340.
Article16. Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. 2017; Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the association for molecular pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 19:4–23. DOI: 10.1016/j.jmoldx.2016.10.002. PMID: 27993330. PMCID: PMC5707196.
Article17. Robinson JT, Thorvaldsdóttir H, Wenger AM, Zehir A, Mesirov JP. 2017; Variant review with the integrative genomics viewer. Cancer Res. 77:e31–4. DOI: 10.1158/0008-5472.CAN-17-0337. PMID: 29092934. PMCID: PMC5678989.
Article18. Jongen-Lavrencic M, Grob T, Hanekamp D, Kavelaars FG, Al Hinai A, Zeilemaker A, et al. 2018; Molecular minimal residual disease in acute myeloid leukemia. N Engl J Med. 378:1189–99. DOI: 10.1056/NEJMoa1716863. PMID: 29601269.
Article19. Yoest JM, Shirai CL, Duncavage EJ. 2020; Sequencing-based measurable residual disease testing in acute myeloid leukemia. Front Cell Dev Biol. 8:249. DOI: 10.3389/fcell.2020.00249. PMID: 32457898. PMCID: PMC7225302.
Article20. Koboldt DC. 2020; Best practices for variant calling in clinical sequencing. Genome Med. 12:91. DOI: 10.1186/s13073-020-00791-w. PMID: 33106175. PMCID: PMC7586657.
Article21. Aynardi J, Manur R, Hess PR, Chekol S, Morrissette JJD, Babushok D, et al. 2018; JAK2 V617F-positive acute myeloid leukaemia (AML): a comparison between de novo AML and secondary AML transformed from an underlying myeloproliferative neoplasm. A study from the Bone Marrow Pathology Group. Br J Haematol. 182:78–85. DOI: 10.1111/bjh.15276. PMID: 29767839.
Article22. Mannelli F. Acute myeloid leukemia evolving from myeloproliferative neoplasms: many sides of a challenging disease. J Clin Med. 2021; 10:DOI: 10.3390/jcm10030436. PMID: 33498691. PMCID: PMC7866045.
Article23. Xia D, Hasserjian RP. 2016; Molecular testing for JAK2, MPL, and CALR in myeloproliferative neoplasms. Am J Hematol. 91:1277–80. DOI: 10.1002/ajh.24578. PMID: 27727468.24. Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, et al. 2007; Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 446:758–64. DOI: 10.1038/nature05690. PMID: 17344859.
Article25. Schwab CJ, Chilton L, Morrison H, Jones L, Al-Shehhi H, Erhorn A, et al. 2013; Genes commonly deleted in childhood B-cell precursor acute lymphoblastic leukemia: association with cytogenetics and clinical features. Haematologica. 98:1081–8. DOI: 10.3324/haematol.2013.085175. PMID: 23508010. PMCID: PMC3696612.
Article26. Thol F, Gabdoulline R, Liebich A, Klement P, Schiller J, Kandziora C, et al. 2018; Measurable residual disease monitoring by NGS before allogeneic hematopoietic cell transplantation in AML. Blood. 132:1703–13. DOI: 10.1182/blood-2018-02-829911. PMID: 30190321. PMCID: PMC7116653.
Article27. Morita K, Kantarjian HM, Wang F, Yan Y, Bueso-Ramos C, Sasaki K, et al. 2018; Clearance of somatic mutations at remission and the risk of relapse in acute myeloid leukemia. J Clin Oncol. 36:1788–97. DOI: 10.1200/JCO.2017.77.6757. PMID: 29702001. PMCID: PMC6008108.
Article28. Kim T, Moon JH, Ahn JS, Kim YK, Lee SS, Ahn SY, et al. 2018; Next-generation sequencing-based posttransplant monitoring of acute myeloid leukemia identifies patients at high risk of relapse. Blood. 132:1604–13. DOI: 10.1182/blood-2018-04-848028. PMID: 30108064.
Article29. Heuser M, Freeman SD, Ossenkoppele GJ, Buccisano F, Hourigan CS, Ngai LL, et al. 2021; 2021 Update on MRD in acute myeloid leukemia: a consensus document from the European LeukemiaNet MRD Working Party. Blood. 138:2753–67. DOI: 10.1182/blood.2021013626. PMID: 34724563. PMCID: PMC8718623.
Article30. Kim HJ, Kim Y, Kang D, Kim HS, Lee JM, Kim M, et al. 2021; Prognostic value of measurable residual disease monitoring by next-generation sequencing before and after allogeneic hematopoietic cell transplantation in acute myeloid leukemia. Blood Cancer J. 11:109. DOI: 10.1038/s41408-021-00500-9. PMID: 34088902. PMCID: PMC8178334.
Article31. Loghavi S, DiNardo CD, Takahashi K, Kanagal-Shamanna R, Tanaka T, Furudate K, et al. 2020; Clonal hematopoiesis and its implications for flow cytometric assessment of measurable residual disease in patients with NPM1-mutated acute myeloid leukemia. Blood. 136:38–9. DOI: 10.1182/blood-2020-139871.32. Klco JM, Miller CA, Griffith M, Petti A, Spencer DH, Ketkar-Kulkarni S, et al. 2015; Association between mutation clearance after induction therapy and outcomes in acute myeloid leukemia. JAMA. 314:811–22. DOI: 10.1001/jama.2015.9643. PMID: 26305651. PMCID: PMC4621257.
Article33. Press RD, Eickelberg G, Froman A, Yang F, Stentz A, Flatley EM, et al. 2019; Next-generation sequencing-defined minimal residual disease before stem cell transplantation predicts acute myeloid leukemia relapse. Am J Hematol. 94:902–12. DOI: 10.1002/ajh.25514. PMID: 31124175.
Article34. Krauth MT, Alpermann T, Bacher U, Eder C, Dicker F, Ulke M, et al. 2015; WT1 mutations are secondary events in AML, show varying frequencies and impact on prognosis between genetic subgroups. Leukemia. 29:660–7. DOI: 10.1038/leu.2014.243. PMID: 25110071.
Article35. Hou HA, Huang TC, Lin LI, Liu CY, Chen CY, Chou WC, et al. 2010; WT1 mutation in 470 adult patients with acute myeloid leukemia: stability during disease evolution and implication of its incorporation into a survival scoring system. Blood. 115:5222–31. DOI: 10.1182/blood-2009-12-259390. PMID: 20368469.36. Bacher U, Haferlach T, Schoch C, Kern W, Schnittger S. 2006; Implications of NRAS mutations in AML: a study of 2502 patients. Blood. 107:3847–53. DOI: 10.1182/blood-2005-08-3522. PMID: 16434492.
Article37. Liu X, Ye Q, Zhao XP, Zhang PB, Li S, Li RQ, et al. 2019; RAS mutations in acute myeloid leukaemia patients: a review and meta-analysis. Clin Chim Acta. 489:254–60. DOI: 10.1016/j.cca.2018.08.040. PMID: 30194935.38. Arber DA, Orazi A, Hasserjian RP, Borowitz MJ, Calvo KR, Kvasnicka HM, et al. 2022; International Consensus Classification of Myeloid neoplasms and Acute Leukemias: integrating morphologic, clinical, and genomic data. Blood. 140:1200–28. DOI: 10.1182/blood.2022015850. PMID: 35767897.39. Khoury JD, Solary E, Abla O, Akkari Y, Alaggio R, Apperley JF, et al. 2022; The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia. 36:1703–19. DOI: 10.1038/s41375-022-01613-1. PMID: 35732831. PMCID: PMC9252913.
Article40. Bean LJH, Funke B, Carlston CM, Gannon JL, Kantarci S, Krock BL, et al. 2020; Diagnostic gene sequencing panels: from design to report-a technical standard of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 22:453–61. DOI: 10.1038/s41436-019-0666-z. PMID: 31732716.
Article41. Yannakou CK, Jones K, McBean M, Thompson ER, Ryland GL, Doig K, et al. 2017; ASXL1 c.1934dup;p.Gly646Trpfs*12-a true somatic alteration requiring a new approach. Blood Cancer J. 7:656. DOI: 10.1038/s41408-017-0025-8. PMID: 29242575. PMCID: PMC5802455.42. Bibault JE, Figeac M, Hélevaut N, Rodriguez C, Quief S, Sebda S, et al. 2015; Next-generation sequencing of FLT3 internal tandem duplications for minimal residual disease monitoring in acute myeloid leukemia. Oncotarget. 6:22812–21. DOI: 10.18632/oncotarget.4333. PMID: 26078355. PMCID: PMC4673201.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Next-Generation Sequencing Myeloid Panel Test for Patients with AML and MDS: Experience in a Tertiary Care Hospital
- Measurable Residual Disease Testing Using Next-Generation Sequencing in Acute Myeloid Leukemia
- Molecular Risk Stratification using Next-generation Sequencing in Acute Myeloid Leukemia
- First Korean Case of Pediatric Acute Megakaryoblastic Leukemia with CBFA2T3::GLIS2 Fusion
- Development of an RNA sequencing panel to detect gene fusions in thyroid cancer