Ann Lab Med.
2023 May;43(3):280-289. 10.3343/alm.2023.43.3.280.
Rapid Targeted Sequencing Using Dried Blood Spot Samples for Patients With Suspected Actionable Genetic Diseases
- Affiliations
-
- 1Department of Genomic Medicine, Seoul National University Hospital, Seoul, Korea
- 2Rare Disease Center, Seoul National University Hospital, Seoul, Korea
- 3Department of Pediatrics, Department of Genome Medicine and Science, Gil Medical Center, Gachon University College of Medicine, Incheon, Korea
- 4MedySapiens, Inc., Seoul, Korea
- 5Department of Electrical Engineering, Gangneung-Wonju National University, Gangneung, Korea
- 6Department of Laboratory Medicine, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea
- 7Department of Pediatrics, Seoul National University Children’s Hospital, Seoul National University College of Medicine, Seoul, Korea
- KMID: 2551712
- DOI: http://doi.org/10.3343/alm.2023.43.3.280
Abstract
- Background
New genome sequencing technologies with enhanced diagnostic efficiency have emerged. Rapid and timely diagnosis of treatable rare genetic diseases can alter their medical management and clinical course. However, multiple factors, including ethical issues, must be considered. We designed a targeted sequencing platform to avoid ethical issues and reduce the turnaround time.
Methods
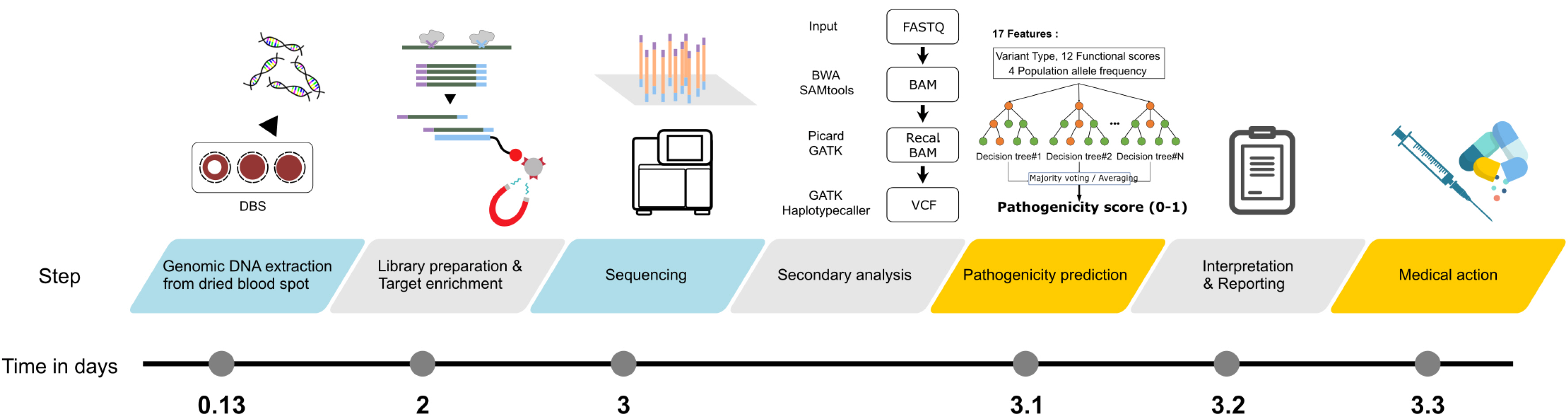
We designed an automated sequencing platform using dried blood spot samples and a NEOseq_ACTION panel comprising 254 genes associated with Mendelian diseases having curable or manageable treatment options. Retrospective validation was performed using data from 24 genetically and biochemically confirmed patients. Prospective validation was performed using data from 111 patients with suspected actionable genetic diseases.
Results
In prospective clinical validation, 13.5% patients presented with medically actionable diseases, including short- or medium-chain acyl-CoA dehydrogenase deficiencies (N=6), hyperphenylalaninemia (N=2), mucopolysaccharidosis type IVA (N=1), alpha thalassemia (N=1), 3-methylcrotonyl-CoA carboxylase 2 deficiency (N=1), propionic acidemia (N=1), glycogen storage disease, type IX(a) (N=1), congenital myasthenic syndrome (N=1), and citrullinemia, type II (N=1). Using the automated analytic pipeline, the turnaround time from blood collection to result reporting was <4 days.
Conclusions
This pilot study evaluated the possibility of rapid and timely diagnosis of treatable rare genetic diseases using a panel designed by a multidisciplinary team. The automated analytic pipeline maximized the clinical utility of rapid targeted sequencing for medically actionable genes, providing a strategy for appropriate and timely treatment of rare genetic diseases.
Keyword
Figure
Cited by 2 articles
-
Rapid Targeted Genomic Testing: A Powerful Tool for Diagnostic Evaluation of Critically Ill Neonates and Infants With Suspected Genetic Diseases
Mi-Ae Jang
Ann Lab Med. 2023;43(3):223-224. doi: 10.3343/alm.2023.43.3.223.Navigating the landscape of clinical genetic testing: insights and challenges in rare disease diagnostics
Soo Yeon Kim
Child Kidney Dis. 2024;28(1):8-15. doi: 10.3339/ckd.24.005.
Reference
-
1. Meng L, Pammi M, Saronwala A, Magoulas P, Ghazi AR, Vetrini F, et al. 2017; Use of exome sequencing for infants in intensive care units: ascertainment of severe single-gene disorders and effect on medical management. JAMA Pediatr. 171:e173438.2. Petrikin JE, Willig LK, Smith LD, Kingsmore SF. 2015; Rapid whole genome sequencing and precision neonatology. Semin Perinatol. 39:623–31. DOI: 10.1053/j.semperi.2015.09.009. PMID: 26521050. PMCID: PMC4657860.
Article3. Willig LK, Petrikin JE, Smith LD, Saunders CJ, Thiffault I, Miller NA, et al. 2015; Whole-genome sequencing for identification of Mendelian disorders in critically ill infants: a retrospective analysis of diagnostic and clinical findings. Lancet Respir Med. 3:377–87. DOI: 10.1016/S2213-2600(15)00139-3. PMID: 25937001.
Article4. Anazi S, Maddirevula S, Faqeih E, Alsedairy H, Alzahrani F, Shamseldin HE, et al. 2017; Clinical genomics expands the morbid genome of intellectual disability and offers a high diagnostic yield. Mol Psychiatry. 22:615–24. DOI: 10.1038/mp.2016.113. PMID: 27431290.
Article5. Stark Z, Tan TY, Chong B, Brett GR, Yap P, Walsh M, et al. 2016; A prospective evaluation of whole-exome sequencing as a first-tier molecular test in infants with suspected monogenic disorders. Genet Med. 18:1090–6. DOI: 10.1038/gim.2016.1. PMID: 26938784.
Article6. Vissers LELM, van Nimwegen KJM, Schieving JH, Kamsteeg EJ, Kleefstra T, Yntema HG, et al. 2017; A clinical utility study of exome sequencing versus conventional genetic testing in pediatric neurology. Genet Med. 19:1055–63. DOI: 10.1038/gim.2017.1. PMID: 28333917. PMCID: PMC5589982.
Article7. Farnaes L, Hildreth A, Sweeney NM, Clark MM, Chowdhury S, Nahas S, et al. 2018; Rapid whole-genome sequencing decreases infant morbidity and cost of hospitalization. NPJ Genom Med. 3:10. DOI: 10.1038/s41525-018-0049-4. PMID: 29644095. PMCID: PMC5884823.
Article8. Stark Z, Lunke S, Brett GR, Tan NB, Stapleton R, Kumble S, et al. 2018; Meeting the challenges of implementing rapid genomic testing in acute pediatric care. Genet Med. 20:1554–63. DOI: 10.1038/gim.2018.37. PMID: 29543227.
Article9. van Diemen CC, Kerstjens-Frederikse WS, Bergman KA, de Koning TJ, Sikkema-Raddatz B, van der Velde JK, et al. 2017; Rapid targeted genomics in critically ill newborns. Pediatrics. 140:e20162854. DOI: 10.1542/peds.2016-2854. PMID: 28939701.
Article10. Gyngell C, Newson AJ, Wilkinson D, Stark Z, Savulescu J. 2019; Rapid challenges: ethics and genomic neonatal intensive care. Pediatrics. 143:S14–21. DOI: 10.1542/peds.2018-1099D. PMID: 30600266. PMCID: PMC6379057.
Article11. Mardis ER. 2010; The $1,000 genome, the $100,000 analysis? Genome Med. 2:84. DOI: 10.1186/gm205. PMID: 21114804. PMCID: PMC3016626.
Article12. Li H, Durbin R. 2009; Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–60. DOI: 10.1093/bioinformatics/btp324. PMID: 19451168. PMCID: PMC2705234.
Article13. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. 2009; The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–9. DOI: 10.1093/bioinformatics/btp352. PMID: 19505943. PMCID: PMC2723002.
Article14. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. 2010; The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–303. DOI: 10.1101/gr.107524.110. PMID: 20644199. PMCID: PMC2928508.
Article15. Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, et al. 2012; A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 6:80–92. DOI: 10.4161/fly.19695. PMID: 22728672. PMCID: PMC3679285.
Article16. Li Q, Wang K. 2017; InterVar: clinical interpretation of genetic variants by the 2015 ACMG-AMP guidelines. Am J Hum Genet. 100:267–80. DOI: 10.1016/j.ajhg.2017.01.004. PMID: 28132688. PMCID: PMC5294755.
Article17. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. 2015; Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 17:405–24. DOI: 10.1038/gim.2015.30. PMID: 25741868. PMCID: PMC4544753.
Article18. Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, et al. 2016; ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 44:D862–8. DOI: 10.1093/nar/gkv1222. PMID: 26582918. PMCID: PMC4702865.
Article19. Stenson PD, Mort M, Ball EV, Chapman M, Evans K, Azevedo L, et al. 2020; The Human Gene Mutation Database (HGMD®): optimizing its use in a clinical diagnostic or research setting. Hum Genet. 139:1197–207. DOI: 10.1007/s00439-020-02199-3. PMID: 32596782. PMCID: PMC7497289.
Article20. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. 2010; A method and server for predicting damaging missense mutations. Nat Methods. 7:248–9. DOI: 10.1038/nmeth0410-248. PMID: 20354512. PMCID: PMC2855889.
Article21. Li B, Krishnan VG, Mort ME, Xin F, Kamati KK, Cooper DN, et al. 2009; Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics. 25:2744–50. DOI: 10.1093/bioinformatics/btp528. PMID: 19734154. PMCID: PMC3140805.
Article22. Kumar P, Henikoff S, Ng PC. 2009; Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 4:1073–81. DOI: 10.1038/nprot.2009.86. PMID: 19561590.
Article23. Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. 2012; Predicting the functional effect of amino acid substitutions and indels. PLoS One. 7:e46688. DOI: 10.1371/journal.pone.0046688. PMID: 23056405. PMCID: PMC3466303.
Article24. Carter H, Douville C, Stenson PD, Cooper DN, Karchin R. 2013; Identifying Mendelian disease genes with the variant effect scoring tool. BMC Genomics. 14(S3):S3. DOI: 10.1186/1471-2164-14-S3-S3. PMID: 23819870. PMCID: PMC3665549.
Article25. Chun S, Fay JC. 2009; Identification of deleterious mutations within three human genomes. Genome Res. 19:1553–61. DOI: 10.1101/gr.092619.109. PMID: 19602639. PMCID: PMC2752137.
Article26. Reva B, Antipin Y, Sander C. 2011; Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 39:e118. DOI: 10.1093/nar/gkr407. PMID: 21727090. PMCID: PMC3177186.
Article27. Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. 2010; MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 7:575–6. DOI: 10.1038/nmeth0810-575. PMID: 20676075.
Article28. Shihab HA, Gough J, Cooper DN, Stenson PD, Barker GL, Edwards KJ, et al. 2013; Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat. 34:57–65. DOI: 10.1002/humu.22225. PMID: 23033316. PMCID: PMC3558800.
Article29. Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, et al. NISC Comparative Sequencing Program. 2005; Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 15:901–13. DOI: 10.1101/gr.3577405. PMID: 15965027. PMCID: PMC1172034.
Article30. Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, et al. 2005; Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15:1034–50. DOI: 10.1101/gr.3715005. PMID: 16024819. PMCID: PMC1182216.
Article31. Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. 2020; The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 581:434–43. DOI: 10.1038/s41586-020-2308-7. PMID: 32461654. PMCID: PMC7334197.
Article32. Karczewski KJ, Weisburd B, Thomas B, Solomonson M, Ruderfer DM, Kavanagh D, et al. 2017; The ExAC browser: displaying reference data information from over 60 000 exomes. Nucleic Acids Res. 45:D840–5. DOI: 10.1093/nar/gkw971. PMID: 27899611. PMCID: PMC5210650.
Article33. Jung KS, Hong KW, Jo HY, Choi J, Ban HJ, Cho SB, et al. 2020; KRGDB: the large-scale variant database of 1722 Koreans based on whole genome sequencing. Database (Oxford). 2020:baz146. DOI: 10.1093/database/baaa030. PMID: 32348452. PMCID: PMC7190023.
Article34. Lee S, Seo J, Park J, Nam JY, Choi A, Ignatius JS, et al. 2017; Korean Variant Archive (KOVA): a reference database of genetic variations in the Korean population. Sci Rep. 7:4287. DOI: 10.1038/s41598-017-04642-4. PMID: 28655895. PMCID: PMC5487339.
Article35. Fu L, Niu B, Zhu Z, Wu S, Li W. 2012; CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 28:3150–2. DOI: 10.1093/bioinformatics/bts565. PMID: 23060610. PMCID: PMC3516142.
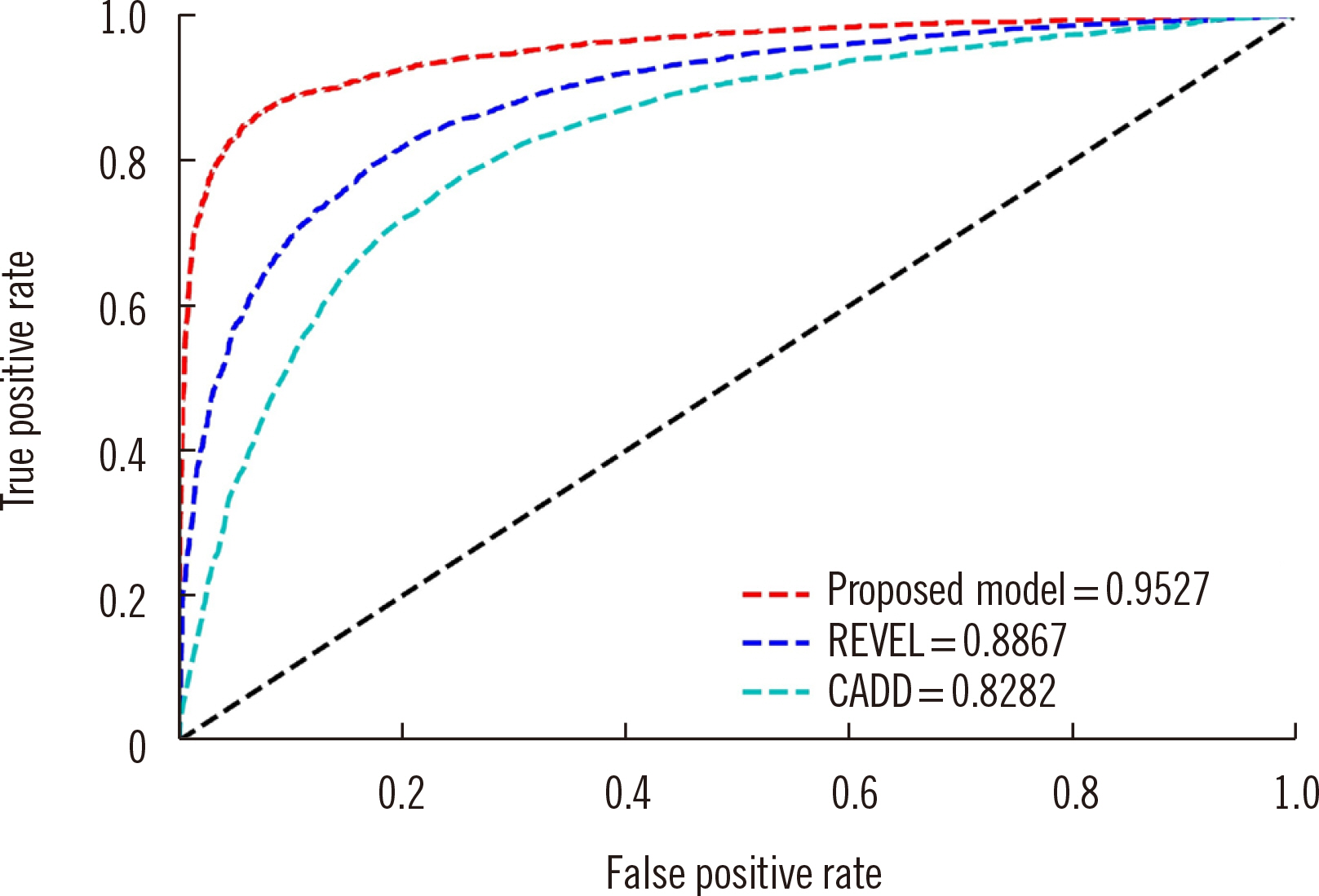
Article36. Ke G, Meng Q, Finley T, Wang T, Chen W, Ma W, et al. 2017; LightGBM: a highly efficient gradient boosting decision tree. Adv Neural Inf Process Syst. 30:3146–54.37. Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, et al. 2016; REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet. 99:877–85. DOI: 10.1016/j.ajhg.2016.08.016. PMID: 27666373. PMCID: PMC5065685.
Article38. Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. 2019; CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 47:D886–94. DOI: 10.1093/nar/gky1016. PMID: 30371827. PMCID: PMC6323892.
Article39. Navarrete R, Leal F, Vega AI, Morais-López A, Garcia-Silva MT, Martín-Hernández E, et al. 2019; Value of genetic analysis for confirming inborn errors of metabolism detected through the Spanish neonatal screening program. Eur J Hum Genet. 27:556–62. DOI: 10.1038/s41431-018-0330-0. PMID: 30626930. PMCID: PMC6460639.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Advances in the use of dried blood spots on filter paper to monitor kidney disease
- Comprehensive Molecular Genetic Analysis in Glioma Patients by Next Generation Sequencing
- A LAMP-SNP Assay Detecting C580Y Mutation in Pfkelch13 Gene from Clinically Dried Blood Spot Samples
- Rapid DNA Extraction from Dried Blood Spots on Filter Paper: Potential Applications in Biobanking
- Concordance of circulating tumor DNA and matched formalin-fixed paraffin-embedded tumor tissue in gastric cancer as a predictor of recurrence