Ann Lab Med.
2023 Mar;43(2):145-152. 10.3343/alm.2023.43.2.145.
Clinical, Mutational, and Transcriptomic Characteristics in Elderly Korean Individuals With Clonal Hematopoiesis Driver Mutations
- Affiliations
-
- 1Divisions of Cardiology, Department of Internal Medicine, Soonchunhyang University Bucheon Hospital, Soonchunhyang University College of Medicine, Bucheon, Korea
- 2Divisions of Hematooncology, Department of Internal Medicine, Soonchunhyang University Bucheon Hospital, Soonchunhyang University College of Medicine, Bucheon, Korea
- 3Department of Laboratory Medicine and Genetics, Soonchunhyang University Bucheon Hospital, Soonchunhyang University College of Medicine, Bucheon, Korea
- KMID: 2551691
- DOI: http://doi.org/10.3343/alm.2023.43.2.145
Abstract
- Background
Clonal hematopoiesis of indeterminate potential (CHIP), which is defined as the presence of blood cells originating from somatically mutated hematopoietic stem cells, is common among the elderly and is associated with an increased risk of hematologic malignancies. We investigated the clinical, mutational, and transcriptomic characteristics in elderly Korean individuals with CHIP mutations.
Methods
We investigated CHIP in 90 elderly individuals aged ≥60 years with normal complete blood counts at a tertiary-care hospital in Korea between June 2021 and February 2022. Clinical and laboratory data were prospectively obtained. Targeted next-generation sequencing of 49 myeloid malignancy driver genes and massively parallel RNA sequencing were performed to explore the molecular spectrum and transcriptomic characteristics of CHIP mutations.
Results
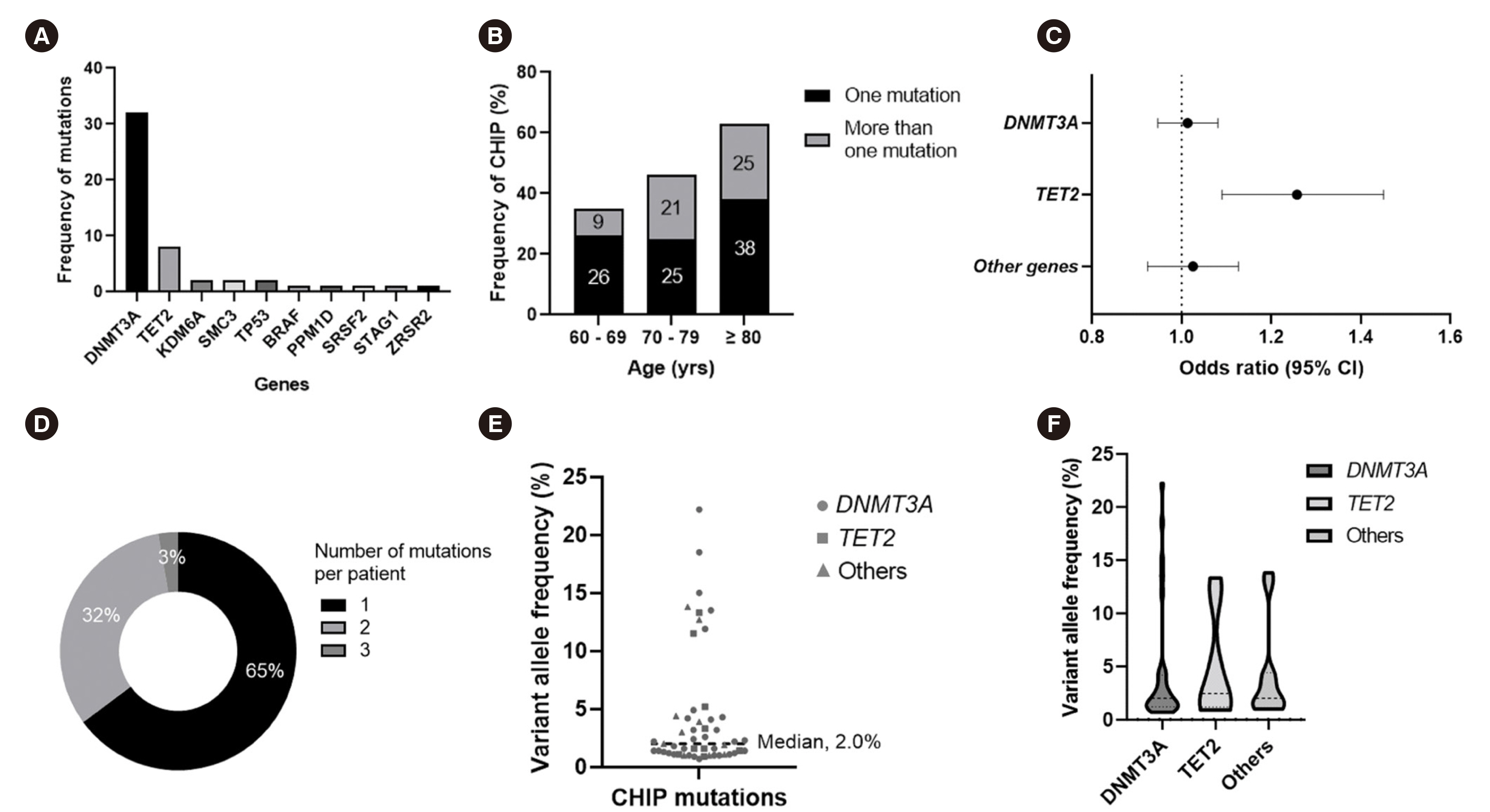
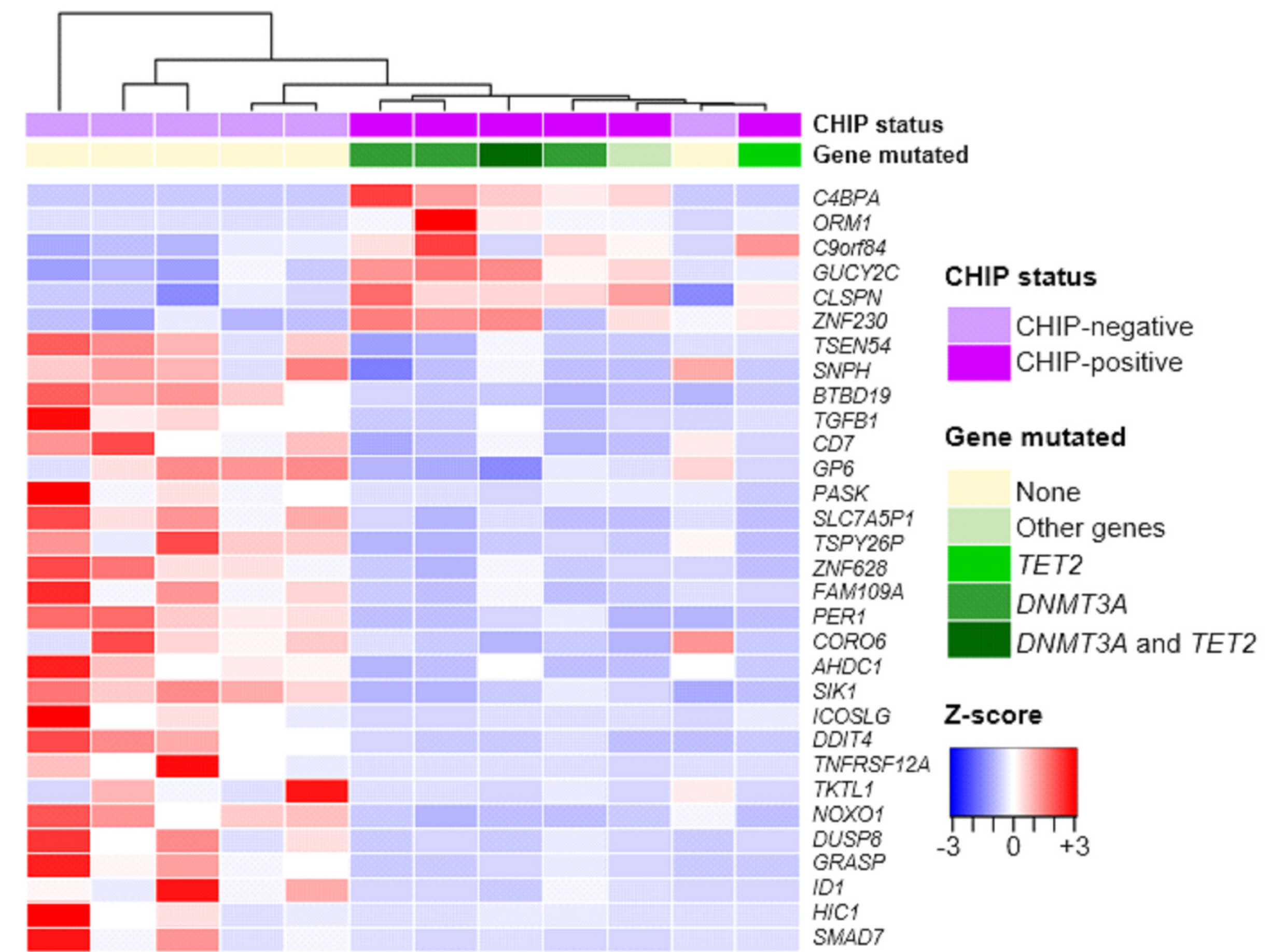
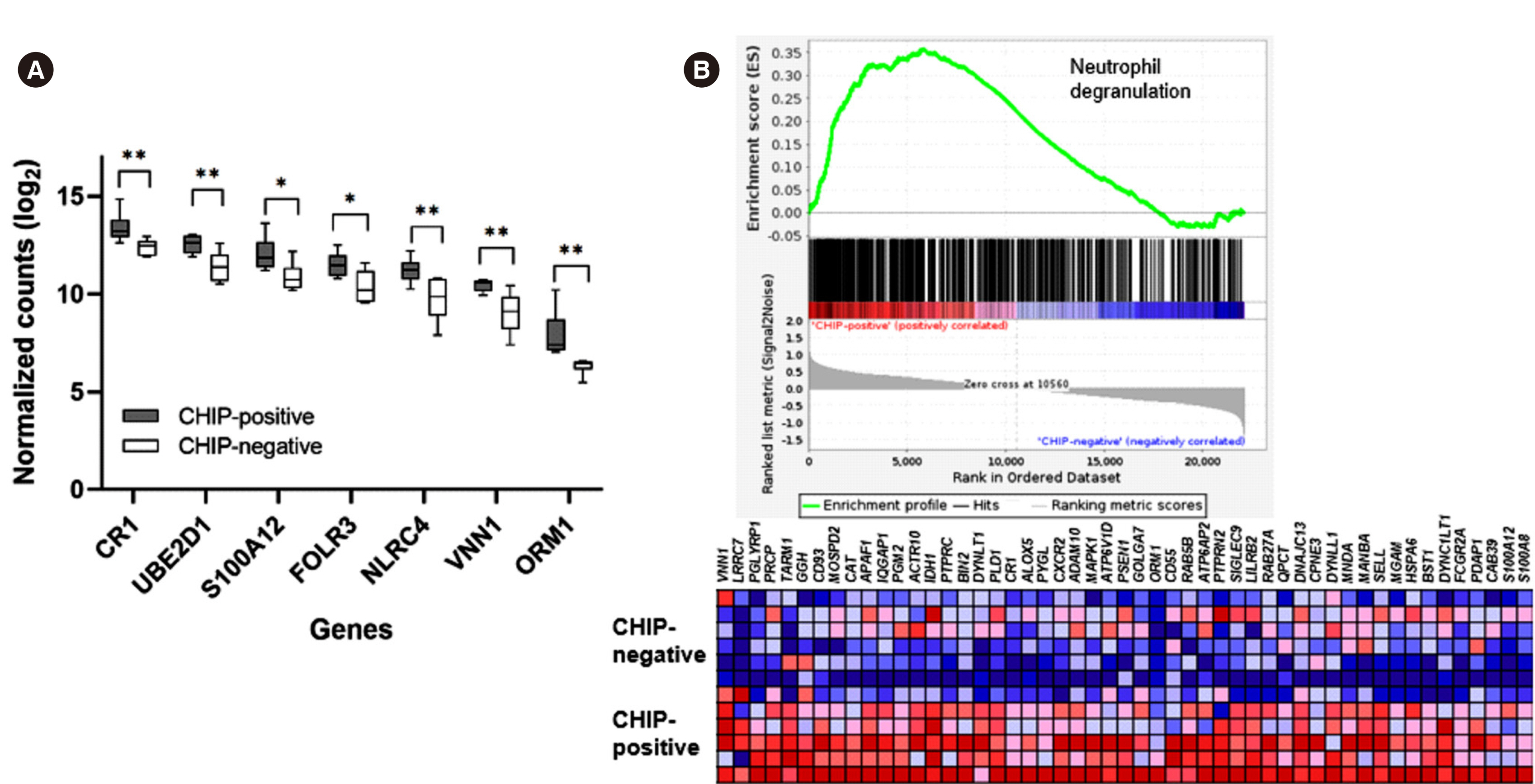
We detected 51 mutations in 10 genes in 37 (41%) of the study individuals. CHIP prevalence increased with age. CHIP mutations were observed with high prevalence in DNMT3A (26 individuals) and TET2 (eight individuals) and were also found in various other genes, including KDM6A, SMC3, TP53, BRAF, PPM1D, SRSF2, STAG1, and ZRSR2. Baseline characteristics, including age, confounding diseases, and blood cell parameters, showed no significant differences. Using mRNA sequencing, we characterized the altered gene expression profile, implicating neutrophil degranulation and innate immune system dysregulation.
Conclusions
Somatic CHIP driver mutations are common among the elderly in Korea and are detected in various genes, including DNMT3A and TET2. Our study highlights that chronic dysregulation of innate immune signaling is associated with the pathogenesis of various diseases, including hematologic malignancies.
Keyword
Figure
Reference
-
1. Busque L, Patel JP, Figueroa ME, Vasanthakumar A, Provost S, Hamilou Z, et al. 2012; Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet. 44:1179–81. DOI: 10.1038/ng.2413. PMID: 23001125. PMCID: PMC3483435.
Article2. Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, et al. 2014; Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 20:1472–8. DOI: 10.1038/nm.3733. PMID: 25326804. PMCID: PMC4313872.
Article3. Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, et al. 2015; Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 126:9–16. DOI: 10.1182/blood-2015-03-631747. PMID: 25931582. PMCID: PMC4624443.
Article4. Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. 2014; Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 371:2488–98. DOI: 10.1056/NEJMoa1408617. PMID: 25426837. PMCID: PMC4306669.
Article5. Evans MA, Sano S, Walsh K. 2020; Cardiovascular disease, aging, and clonal hematopoiesis. Annu Rev Pathol. 15:419–38. DOI: 10.1146/annurev-pathmechdis-012419-032544. PMID: 31689371. PMCID: PMC7104598.
Article6. Jaiswal S. 2020; Clonal hematopoiesis and nonhematologic disorders. Blood. 136:1606–14. DOI: 10.1182/blood.2019000989. PMID: 32736379. PMCID: PMC8209629.
Article7. Leoni C, Montagner S, Rinaldi A, Bertoni F, Polletti S, Balestrieri C, et al. 2017; Dnmt3a restrains mast cell inflammatory responses. Proc Natl Acad Sci U S A. 114:E1490–9. DOI: 10.1073/pnas.1616420114. PMID: 28167789. PMCID: PMC5338402.8. Zhang Q, Zhao K, Shen Q, Han Y, Gu Y, Li X, et al. 2015; Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature. 525:389–93. DOI: 10.1038/nature15252. PMID: 26287468. PMCID: PMC4697747.
Article9. Jaiswal S, Ebert BL. 2019; Clonal hematopoiesis in human aging and disease. Science. 366:eaan4673. DOI: 10.1126/science.aan4673. PMID: 31672865. PMCID: PMC8050831.
Article10. Zhong Y, Xu F, Wu J, Schubert J, Li MM. 2021; Application of next generation sequencing in laboratory medicine. Ann Lab Med. 41:25–43. DOI: 10.3343/alm.2021.41.1.25. PMID: 32829577. PMCID: PMC7443516.
Article11. Park HS, Son SM, Kwon J. 2022; Serial analysis and comparison of mutation profiles in decitabine-treated myeloid sarcoma and subsequent acute myeloid leukemia using next-generation sequencing. Ann Lab Med. 42:602–5. DOI: 10.3343/alm.2022.42.5.602. PMID: 35470279. PMCID: PMC9057818.
Article12. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. 2013; STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. DOI: 10.1093/bioinformatics/bts635. PMID: 23104886. PMCID: PMC3530905.
Article13. Love MI, Huber W, Anders S. 2014; Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. DOI: 10.1186/s13059-014-0550-8. PMID: 25516281. PMCID: PMC4302049.
Article14. Mi H, Ebert D, Muruganujan A, Mills C, Albou LP, Mushayamaha T, et al. 2021; PANTHER version 16: a revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 49:D394–403. DOI: 10.1093/nar/gkaa1106. PMID: 33290554. PMCID: PMC7778891.
Article15. Fabregat A, Sidiropoulos K, Viteri G, Marin-Garcia P, Ping P, Stein L, et al. 2018; Reactome diagram viewer: data structures and strategies to boost performance. Bioinformatics. 34:1208–14. DOI: 10.1093/bioinformatics/btx752. PMID: 29186351. PMCID: PMC6030826.
Article16. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. 2005; Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 102:15545–50. DOI: 10.1073/pnas.0506580102. PMID: 16199517. PMCID: PMC1239896.
Article17. Cuartero S, Innes AJ, Merkenschlager M. 2019; Towards a better understanding of cohesin mutations in AML. Front Oncol. 9:867. DOI: 10.3389/fonc.2019.00867. PMID: 31552185. PMCID: PMC6746210.
Article18. Viny AD, Bowman RL, Liu Y, Lavallée VP, Eisman SE, Xiao W, et al. 2019; Cohesin members Stag1 and Stag2 display distinct roles in chromatin accessibility and topological control of HSC Self-renewal and differentiation. Cell Stem Cell. 25:682–96.e8. DOI: 10.1016/j.stem.2019.08.003. PMID: 31495782. PMCID: PMC6842438.
Article19. Cuartero S, Weiss FD, Dharmalingam G, Guo Y, Ing-Simmons E, Masella S, et al. 2018; Control of inducible gene expression links cohesin to hematopoietic progenitor self-renewal and differentiation. Nat Immunol. 19:932–41. DOI: 10.1038/s41590-018-0184-1. PMID: 30127433. PMCID: PMC6195188.
Article20. Thota S, Viny AD, Makishima H, Spitzer B, Radivoyevitch T, Przychodzen B, et al. 2014; Genetic alterations of the cohesin complex genes in myeloid malignancies. Blood. 124:1790–8. DOI: 10.1182/blood-2014-04-567057. PMID: 25006131. PMCID: PMC4162108.
Article21. Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, et al. Cancer Genome Atlas Research Network. 2013; Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 368:2059–74. DOI: 10.1056/NEJMoa1301689. PMID: 23634996. PMCID: PMC3767041.
Article22. Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, et al. 2012; The origin and evolution of mutations in acute myeloid leukemia. Cell. 150:264–78. DOI: 10.1016/j.cell.2012.06.023. PMID: 22817890. PMCID: PMC3407563.
Article23. Thol F, Bollin R, Gehlhaar M, Walter C, Dugas M, Suchanek KJ, et al. 2014; Mutations in the cohesin complex in acute myeloid leukemia: clinical and prognostic implications. Blood. 123:914–20. DOI: 10.1182/blood-2013-07-518746. PMID: 24335498.
Article24. Barreyro L, Chlon TM, Starczynowski DT. 2018; Chronic immune response dysregulation in MDS pathogenesis. Blood. 132:1553–60. DOI: 10.1182/blood-2018-03-784116. PMID: 30104218. PMCID: PMC6182269.
Article25. Mantovani A, Cassatella MA, Costantini C, Jaillon S. 2011; Neutrophils in the activation and regulation of innate and adaptive immunity. Nat Rev Immunol. 11:519–31. DOI: 10.1038/nri3024. PMID: 21785456.
Article26. Rawat K, Syeda S, Shrivastava A. 2021; Neutrophil-derived granule cargoes: paving the way for tumor growth and progression. Cancer Metastasis Rev. 40:221–44. DOI: 10.1007/s10555-020-09951-1. PMID: 33438104. PMCID: PMC7802614.
Article27. Filep JG. 2022; Targeting neutrophils for promoting the resolution of inflammation. Front Immunol. 13:866747. DOI: 10.3389/fimmu.2022.866747. PMID: 35371088. PMCID: PMC8966391.
Article28. Liew PX, Kubes P. 2019; The neutrophil's role during health and disease. Physiol Rev. 99:1223–48. DOI: 10.1152/physrev.00012.2018. PMID: 30758246.
Article29. Jaiswal S, Natarajan P, Silver AJ, Gibson CJ, Bick AG, Shvartz E, et al. 2017; Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med. 377:111–21. DOI: 10.1056/NEJMoa1701719. PMID: 28636844. PMCID: PMC6717509.
Article30. Kiefer KC, Cremer S, Pardali E, Assmus B, Abou-El-Ardat K, Kirschbaum K, et al. 2021; Full spectrum of clonal haematopoiesis-driver mutations in chronic heart failure and their associations with mortality. ESC Heart Fail. 8:1873–84. DOI: 10.1002/ehf2.13297. PMID: 33779075. PMCID: PMC8120376.
Article31. van Zeventer IA, de Graaf AO, Wouters HJCM, van der Reijden BA, van der Klauw MM, de Witte T, et al. 2020; Mutational spectrum and dynamics of clonal hematopoiesis in anemia of older individuals. Blood. 135:1161–70. DOI: 10.1182/blood.2019004362. PMID: 32243522.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Current Aspects of Clonal Hematopoiesis: Implications for Clinical Diagnosis
- Clinical Significance of Clonal Hematopoiesis
- Clonal hematopoiesis: elements associated with clonal expansion and diseases
- A Rare Case of Essential Thrombocythemia with Coexisting JAK2 and MPL Driver Mutations
- Association Between Clonal Hematopoiesis of Indeterminate Potential and Brain β-Amyloid Deposition in Korean Patients With Cognitive Impairment