Ann Clin Microbiol.
2023 Dec;26(4):117-124. 10.5145/ACM.2023.26.4.117.
Evaluation of the efficacy of three medical device detergents on bacteria and yeast derived biofilm: a comparative study
- Affiliations
-
- 1Department of Laboratory Medicine, Sanggye Paik Hospital, Inje University College of Medicine, Seoul, Korea
- 2Department of Laboratory Medicine, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea
- KMID: 2550663
- DOI: http://doi.org/10.5145/ACM.2023.26.4.117
Abstract
- Background
This study aimed to evaluate the efficacy of three medical detergents against bacteria and yeast-derived biofilms.
Methods
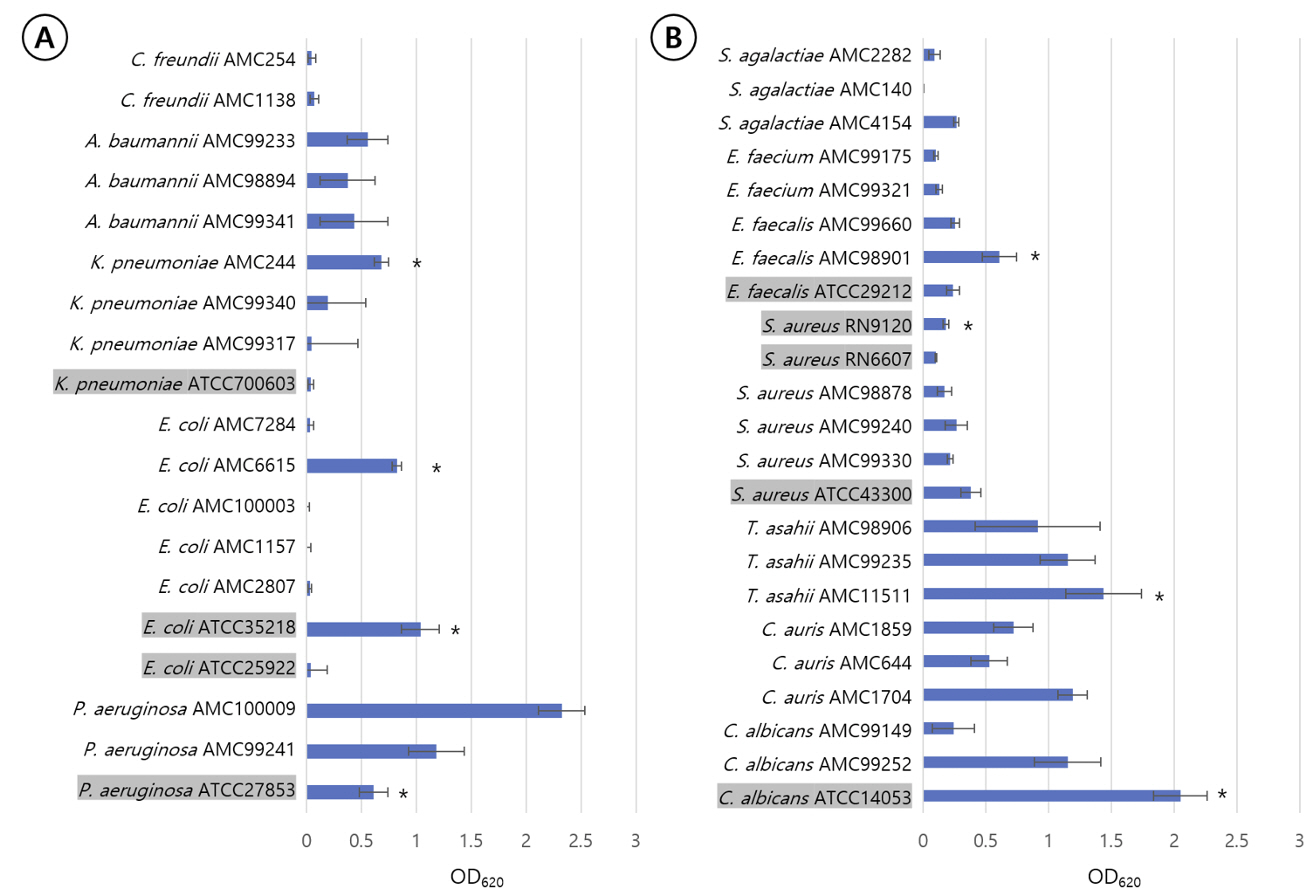
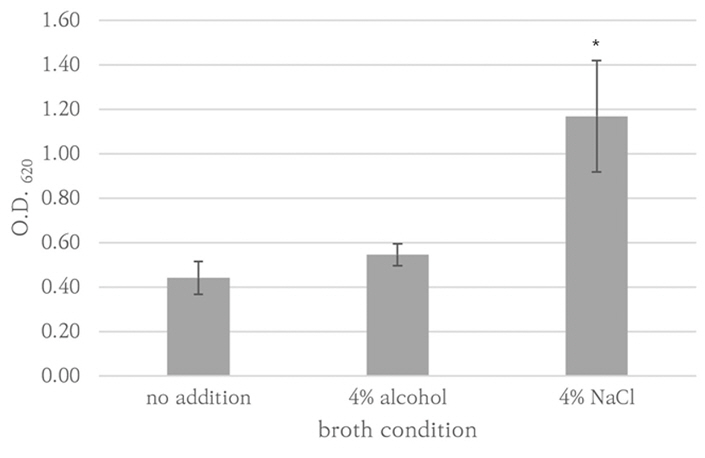
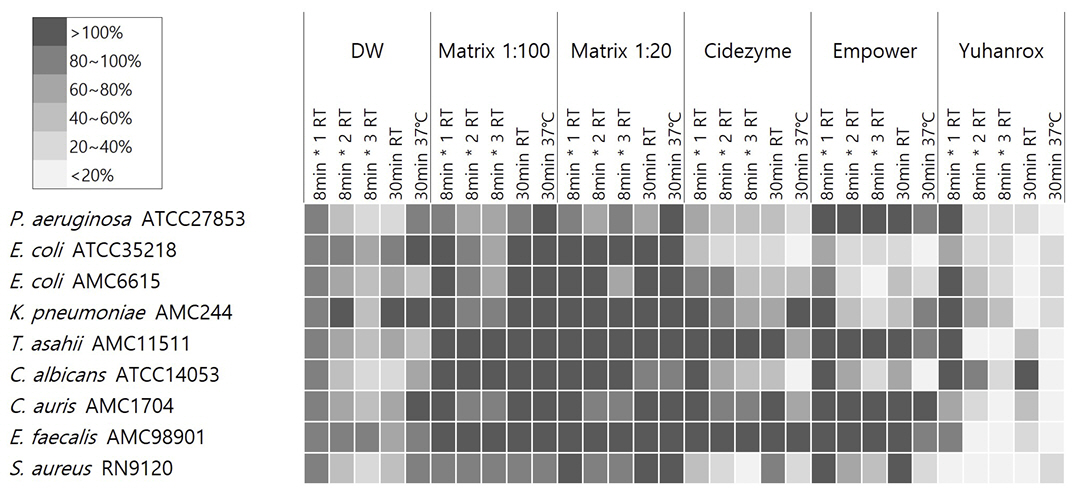
The biofilm removal efficacy of Empower TM (Metrex, USA), Cidezyme TM (Johnson and Johnson Medical Inc, USA), and Matrix mint TM (Whiteley Medical, Australia) were compared to that of chlorine bleach. Biofilms were produced using Staphylococcus aureusRN9120, Escherichia coli ATCC35218, Pseudomonas aeruginosa ATCC27853, Candida albicans ATCC14053, and clinical isolates of Enterococcus faecalis, E. coli, Klebsiella pneumoniae, Candida auris, and Trichosporon asahii. The organisms were suspended in tryptic soy broth (TSB) in 96-well microplates and cultured for 72 hours. They were treated with the detergents, and the residual biofilm mass was quantified using crystal violet staining followed by optical density measurements at 620 nm (OD 620 ).
Results
Empower TM and Cidezyme TM significantly reduced the biofilm mass derived from all species by > 50% of OD 620 at 37ºC except those from E. faecalis, T. asahii, and C. auris. Matrix mint TM had no effect on the biofilms under any condition.
Conclusion
The culture conditions and the species of the biofilm-producing organism influenced the effectiveness of the detergent. Biofilms produced by E. faecalis, C. auris, and T.asahii were resistant to all detergent treatments under all conditions.
Figure
Reference
-
1. Davies D. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov 2003;2:114-22. .2. Sutherland IW. The biofilm matrix-an immobilized but dynamic microbial environment. Trends Microbiol 2001;9:222-7. .3. Flemming HC and Wingender J. Relevance of microbial extracellular polymeric substances (EPSs)--part I: structural and ecological aspects. Water Sci Technol 2001;43:1-8. .4. Centers for Disease Control and Prevention. Guideline for disinfection and sterilization in healthcare facilities (2008). https://www.cdc.gov/infectioncontrol/guidelines/disinfection/ index.html [Online] (last visited on 1 Septebmer 2023). .5. Wang S, Zhao Y, Breslawec AP, Liang T, Deng Z, Kuperman LL, et al. Strategy to combat biofilms: a focus on biofilm dispersal enzymes. NPJ Biofilms Microbiomes 2023;9:63. .6. Wilcox MH, Fawley WN, Wigglesworth N, Parnell P, Verity P, Freeman J. Comparison of the effect of detergent versus hypochlorite cleaning on environmental contamination and incidence of Clostridium dif f icile infection. J Hosp Infect 2003;54:109-14. .7. Gonzalez JA, Vanzieleghem T, Dumazy A, Meuris C, Mutsers J, Christiaens G, et al. On-site comparison of an enzymatic detergent and a non-enzymatic detergent-disinfectant for routine manual cleaning of flexible endoscopes. Endosc Int Open 2019;7:E412-20. .8. Zühlsdorf B, Emmrich M, Floss H, Martiny H. Cleaning efficacy of nine different cleaners in a washer–disinfector designed for flexible endoscopes. J Hosp Infect 2002;52:206-11. .9. Fang Y, Shen Z, Li L, Cao Y, Gu LY, Gu Q, et al. A study of the efficacy of bacterial biofilm cleanout for gastrointestinal endoscopes. World J Gastroenterol 2010;16:1019-24. .10. Ren W, Sheng X, Huang X, Zhi F, Cai W. Evaluation of detergents and contact time on biofilm removal from flexible endoscopes. Am J Infect Control 2013;41:e89-92. .11. Vickery K, Pajkos A, Cossart Y. Removal of biofilm from endoscopes: evaluation of detergent efficiency. Am J Infect Control 2004;32:170-6. .12. da Costa Luciano C, Olson N, Tipple AF, Alfa M. Evaluation of the ability of different detergents and disinfectants to remove and kill organisms in traditional biofilm. Am J Infect Control 2016;44:e243-9. .13. Sakoulas G, Eliopoulos GM, Moellering RC, Jr., Wennersten C, Venkataraman L, Novick RP, et al. Accessory gene regulator (agr) locus in geographically diverse Staphylococcus aureus isolates with reduced susceptibility to vancomycin. Antimicrob Agents Chemother 2002;46:1492-502. .14. Mangwani N, Shukla SK, Kumari S, Das S, Rao TS. Effect of biofilm parameters and extracellular polymeric substance composition on polycyclic aromatic hydrocarbon degradation. RSC Adv 2016;6:S7540-51. .15. Musleh RM and Jebur AQ. Crystal violet binding assay for assessment of biofilm formation by Klebsiella pneumoniae on catheter, glass and stainless-steel surfaces. Iraqi J Sci 2014;55:120812. .16. Stiefel P, Mauerhofer S, Schneider J, Maniura-Weber K, Rosenberg U, Ren Q. Enzymes enhance biofilm removal efficiency of cleaners. Antimicrob Agents Chemother 2016;60:364752. .17. Stiefel P, Rosenberg U, Schneider J, Mauerhofer S, Maniura-Weber K, Ren Q. Is biofilm removal properly assessed? Comparison of different quantification methods in a 96-well plate system. Appl Microbiol Biotechnol 2016;100:4135-45. .18. Lee S, Choi KH, Yoon Y. Effect of NaCl on biofilm formation of the isolate from Staphylococcus aureus outbreak linked to ham. Korean J Food Sci Anim Resour 2014;34:25761. .19. Borucki MK, Peppin JD, White D, Loge F, Call DR. Variation in biofilm formation among strains of Listeria monocytogenes. Appl Environ Microbiol 2003;69:7336-42. .20. Lianou A, Nychas GE, Koutsoumanis KP. Strain variability in biofilm formation: a food safety and quality perspective. Food Res Int 2020;137:109424. .21. Hoiby N, Bjarnsholt T, Moser C, Bassi GL, Coenye T, Donelli G, et al. ESCMID guideline for the diagnosis and treatment of biofilm infections 2014. Clin Microbiol Infect 2015;21:S1-25. .22. Fanning S and Mitchell AP. Fungal biofilms. PLoS Pathog 2012;8:e1002585. .23. Rahman MR, Perisetti A, Coman R, Bansal P, Chhabra R, Goyal H. Duodenoscope-associated infections: update on an emerging problem. Dig Dis Sci 2019;64:1409-18. .24. Aumeran C, Poincloux L, Souweine B, Robin F, Laurichesse H, Baud O, et al. Multidrug-resistant Klebsiella pneumoniae outbreak after endoscopic retrograde cholangiopancreatography. Endoscopy 2010;42:895-9. .25. Molobela IP, Cloete TE, Beukes M. Protease and amylase enzymes for biofilm removal and degradation of extracellular polymeric substances (EPS) produced by Pseudomonas f luorescens bacteria. Afr J Microbiol Res 2010;4:1515-24. .26. Tristezza M, Lourenço A, Barata A, Brito L, Malfeito-Ferreira M, Loureiro V. Susceptibility of wine spoilage yeasts and bacteria in the planktonic state and in biofilms to disinfectants. Ann Microbiol 2010;60:549-56. .27. Sun W, Wang Y, Zhang W, Ying H, Wang P. Novel surfactant peptide for removal of biofilms. Colloids Surf B Biointerfaces 2018;172:180-6. .28. Ribeiro MM, Graziano KU, Olson N, França R, Alfa MJ. The polytetrafluoroethylene (PTFE) channel model of cyclic-buildup biofilm and traditional biofilm: the impact of friction, and detergent on cleaning and subsequent high-level disinfection. Infect Control Hosp Epidemiol 2020;41:172-80. .29. Beilenhoff U, Biering H, Blum R, Brljak J, Cimbro M, Dumonceau JM, et al. Reprocessing of f lexible endoscopes and endoscopic accessories used in gastrointestinal endoscopy: position statement of the European Society of Gastrointestinal Endoscopy (ESGE) and European Society of Gastroenterology Nurses and Associates (ESGENA) - update 2018. Endoscopy 2018;50:1205-34.30. Maillard JY and Centeleghe I. How biofilm changes our understanding of cleaning and disinfection. Antimicrob Resist Infect Control 2023;12:95. .31. Wolcott R, Costerton JW, Raoult D, Cutler SJ. The polymicrobial nature of biofilm infection. Clin Microbiol Infect 2013;19:107-12. .
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Influence of Bacterial Presence on Biofilm Formation of Candida albicans
- Biofilm
- Inhibitory Effect of Pentose on Biofilm Formation by Oral Bacteria
- Preventive effects of shiitake mushroom extract on candida stomatitis
- Effects of Biofilm Formation on The Antimicrobial Susceptibility of Staphylococcus aureus