Cancer Res Treat.
2023 Oct;55(4):1303-1312. 10.4143/crt.2023.291.
Prevalence and Risk Factors of Germline Pathogenic Variants in Pancreatic Ductal Adenocarcinoma
- Affiliations
-
- 1Center for Cancer Prevention and Detection, National Cancer Center, Goyang, Korea
- 2Targeted Therapy Branch, Center for Rare Cancers, National Cancer Center, Goyang, Korea
- 3Center for Liver and Pancreatobiliary Cancer, National Cancer Center, Goyang, Korea
- 4GC Genome, Green Cross Laboratories, Yongin, Korea
- 5Biostatics Collaboration Team, Research Institute, National Cancer Center, Goyang, Korea
- 6Department of Laboratory Medicine, Hospital, National Cancer Center, Goyang, Korea
- 7Cancer Biomedical Science, National Cancer Center Graduate School of Cancer Science and Policy, Goyang, Korea
- 8Division of Convergence Technology, Research Institute, National Cancer Center, Goyang, Korea
- KMID: 2547804
- DOI: http://doi.org/10.4143/crt.2023.291
Abstract
- Purpose
The genetic attribution for pancreatic ductal adenocarcinoma (PDAC) has been reported as 5%-10%. However, the incidence of germline pathogenic variants (PVs) in Korean PDAC patients has not been thoroughly investigated. Therefore, we studied to identify the risk factors and prevalence of PV for future treatment strategies in PDAC.
Materials and Methods
Total of 300 (155 male) patients with a median age of 65 years (range, 33 to 90 years) were enrolled in National Cancer Center in Korea. Cancer predisposition genes, clinicopathologic characteristics, and family history of cancer were analyzed.
Results
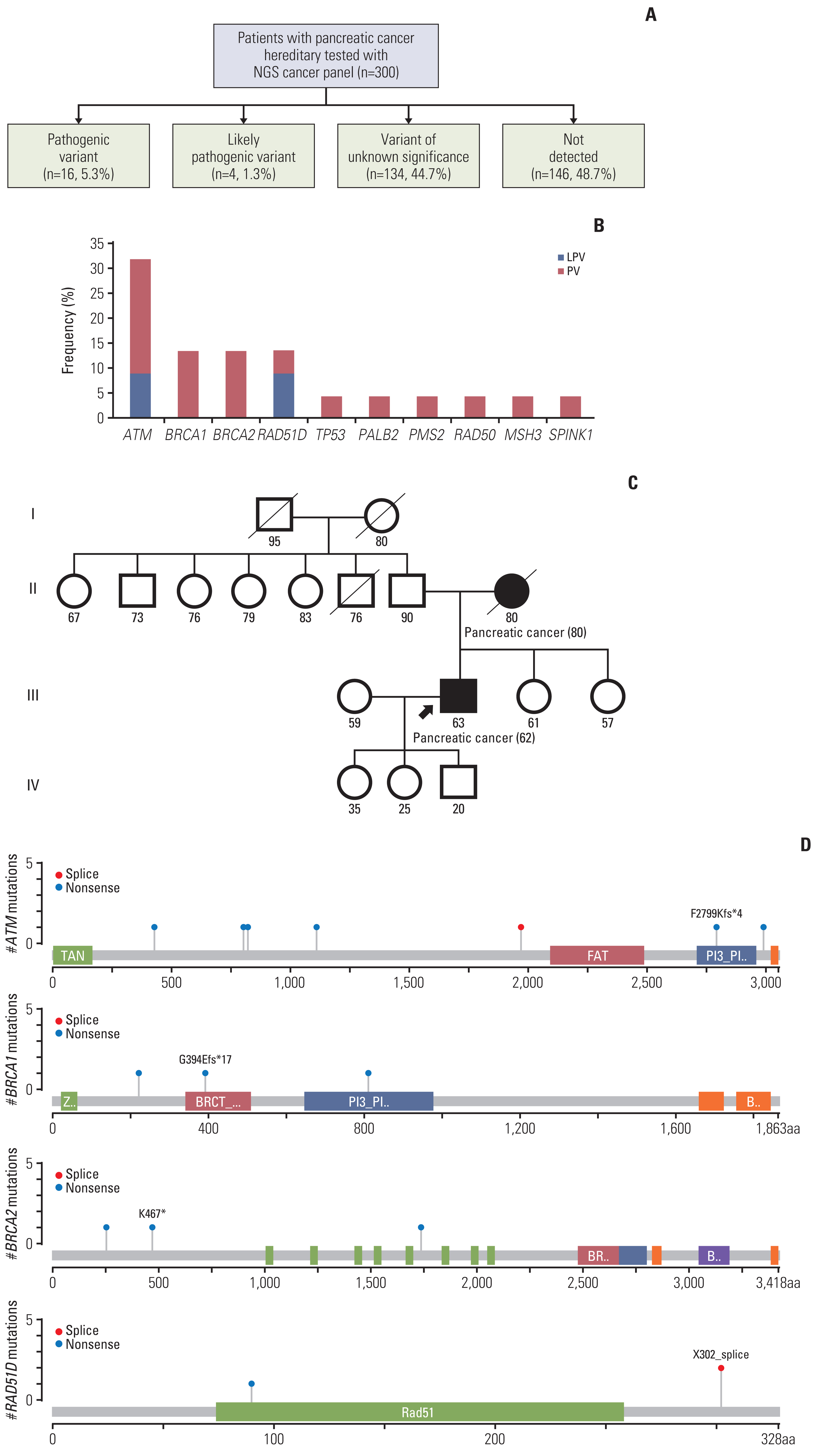
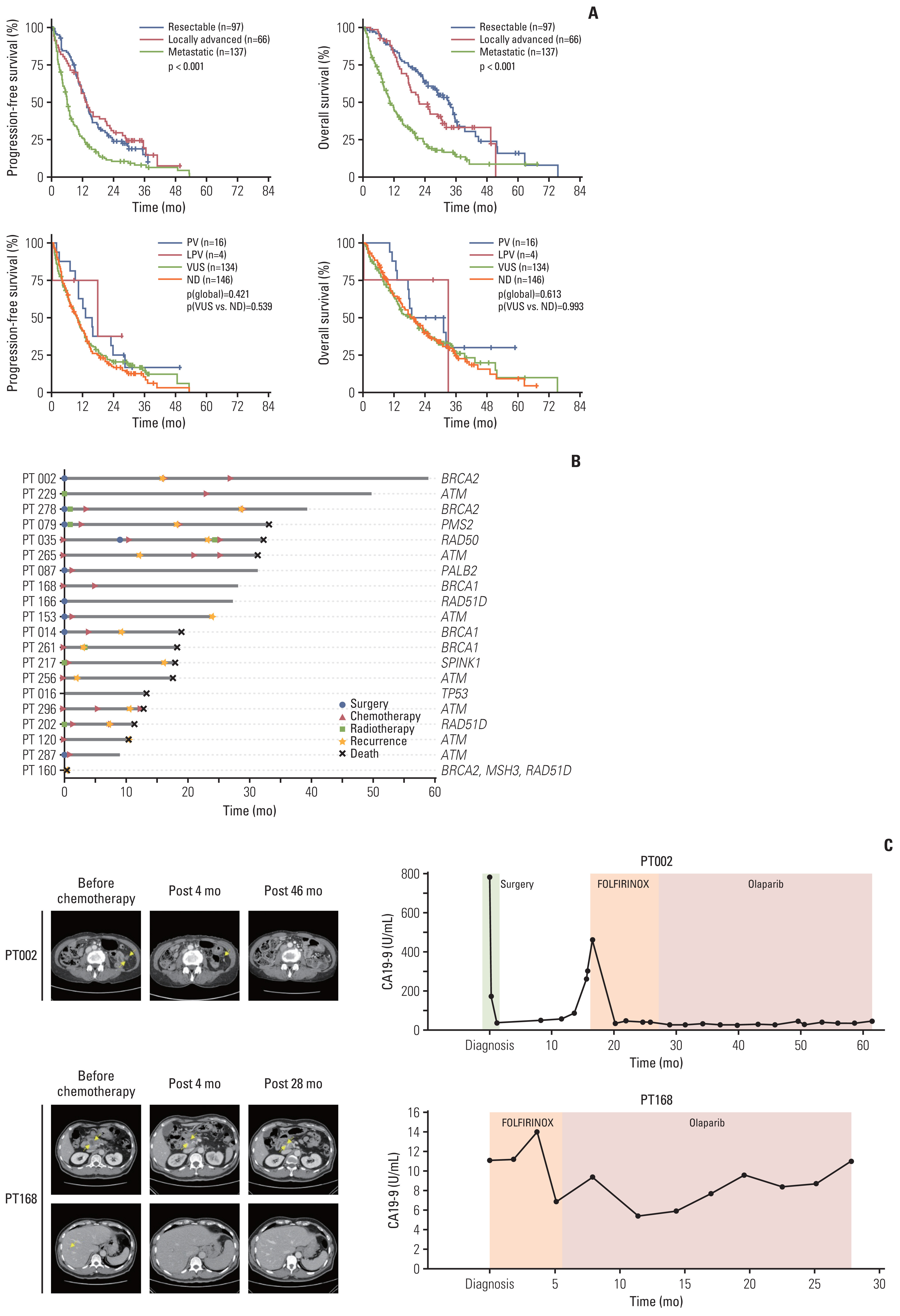
PVs were detected in 20 patients (6.7%, median age 65) in ATM (n=7, 31.8%), BRCA1 (n=3, 13.6%), BRCA2 (n=3), and RAD51D (n=3). Each one patient showed TP53, PALB2, PMS2, RAD50, MSH3, and SPINK1 PV. Among them, two likely PVs were in ATM and RAD51D, respectively. Family history of various types of cancer including pancreatic cancer (n=4) were found in 12 patients. Three patients with ATM PVs and a patient with three germline PVs (BRCA2, MSH3, and RAD51D) had first-degree relatives with pancreatic cancer. Familial pancreatic cancer history and PVs detection had a significant association (4/20, 20% vs. 16/264, 5.7%; p=0.035).
Conclusion
Our study demonstrated that germline PVs in ATM, BRCA1, BRCA2, and RAD51D are most frequent in Korean PDAC patients and it is comparable to those of different ethnic groups. Although this study did not show guidelines for germline predisposition gene testing in patients with PDAC in Korea, it would be emphasized the need for germline testing for all PDAC patients.
Figure
Reference
-
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021; 71:209–49.2. National Cancer Information Center [Internet]. Goyang: Natioanl Cancer Information Center;2019. [cited 2023 Mar 31]. Available from: https://www.cancer.go.kr/ .3. Tempero MA, Malafa MP, Al-Hawary M, Behrman SW, Benson AB, Cardin DB, et al. Pancreatic adenocarcinoma, version 2.2021, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2021; 19:439–57.4. Abe K, Kitago M, Kitagawa Y, Hirasawa A. Hereditary pancreatic cancer. Int J Clin Oncol. 2021; 26:1784–92.5. Okur V, Chung WK. The impact of hereditary cancer gene panels on clinical care and lessons learned. Cold Spring Harb Mol Case Stud. 2017; 3:a002154.6. Tempero MA. NCCN guidelines updates: pancreatic cancer. J Natl Compr Canc Netw. 2019; 17:603–5.7. Daly MB, Pal T, Berry MP, Buys SS, Dickson P, Domchek SM, et al. Genetic/familial high-risk assessment: breast, ovarian, and pancreatic, version 2.2021, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2021; 19:77–102.8. Astiazaran-Symonds E, Goldstein AM. A systematic review of the prevalence of germline pathogenic variants in patients with pancreatic cancer. J Gastroenterol. 2021; 56:713–21.9. DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011; 43:491–8.10. Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at: https://arxiv.org/abs/1303.3997 (2013).11. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010; 20:1297–303.12. Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011; 27:2987–93.13. Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. Preprint at: https://arxiv.org/abs/1207.3907 (2012).14. McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, et al. The Ensembl Variant Effect Predictor. Genome Biol. 2016; 17:122.15. Liu X, Wu C, Li C, Boerwinkle E. dbNSFP v3.0: a one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum Mutat. 2016; 37:235–41.16. cBioPortal for Cancer Genomics [Internet]. cBioPortal; 2022 [cited 2023 Mar 31]. Available from: http://www.cbioportal.org.17. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015; 17:405–24.18. Hu C, Hart SN, Bamlet WR, Moore RM, Nandakumar K, Eckloff BW, et al. Prevalence of pathogenic mutations in cancer predisposition genes among pancreatic cancer patients. Cancer Epidemiol Biomarkers Prev. 2016; 25:207–11.19. Hu C, Hart SN, Polley EC, Gnanaolivu R, Shimelis H, Lee KY, et al. Association between inherited germline mutations in cancer predisposition genes and risk of pancreatic cancer. JAMA. 2018; 319:2401–9.20. Shindo K, Yu J, Suenaga M, Fesharakizadeh S, Cho C, Macgregor-Das A, et al. Deleterious germline mutations in patients with apparently sporadic pancreatic adenocarcinoma. J Clin Oncol. 2017; 35:3382–90.21. Chaffee KG, Oberg AL, McWilliams RR, Majithia N, Allen BA, Kidd J, et al. Prevalence of germ-line mutations in cancer genes among pancreatic cancer patients with a positive family history. Genet Med. 2018; 20:119–27.22. Yurgelun MB, Chittenden AB, Morales-Oyarvide V, Rubinson DA, Dunne RF, Kozak MM, et al. Germline cancer susceptibility gene variants, somatic second hits, and survival outcomes in patients with resected pancreatic cancer. Genet Med. 2019; 21:213–23.23. Earl J, Galindo-Pumarino C, Encinas J, Barreto E, Castillo ME, Pachon V, et al. A comprehensive analysis of candidate genes in familial pancreatic cancer families reveals a high frequency of potentially pathogenic germline variants. EBioMedicine. 2020; 53:102675.24. Wieme G, Kral J, Rosseel T, Zemankova P, Parton B, Vocka M, et al. Prevalence of germline pathogenic variants in cancer predisposing genes in Czech and Belgian pancreatic cancer patients. Cancers (Basel). 2021; 13:4430.25. Lee K, Yoo C, Kim KP, Park KJ, Chang HM, Kim TW, et al. Germline BRCA mutations in Asian patients with pancreatic adenocarcinoma: a prospective study evaluating risk category for genetic testing. Invest New Drugs. 2018; 36:163–9.26. Park JH, Jo JH, Jang SI, Chung MJ, Park JY, Bang S, et al. BRCA 1/2 germline mutation predicts the treatment response of FOLFIRINOX with pancreatic ductal adenocarcinoma in Korean patients. Cancers (Basel). 2022; 14:236.27. National Health Insurance Act. Approval from Ministry of Health and Welfare (Notice 2020-135) (2020 Jul 1).28. Vidula N, Rich TA, Sartor O, Yen J, Hardin A, Nance T, et al. Routine plasma-based genotyping to comprehensively detect germline, somatic, and reversion BRCA mutations among patients with advanced solid tumors. Clin Cancer Res. 2020; 26:2546–55.29. Walker EJ, Blanco AM, Carnevale J, Cinar P, Collisson EA, Tempero MA, et al. The additional diagnostic benefit of pancreatic cancer molecular profiling after germline testing. Pancreas. 2022; 51:302–4.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Germline pathogenic variants in unselected Korean men with prostate cancer
- Pathological Classification of Panaeatic Cancer and Precancerous Casion
- A Serous Cystic Neoplasm of the Pancreas with Synchronous Pancreatic Ductal Adenocarcinoma
- Founder BRCA1 mutations in Nepalese population
- Exploring the variations of the pancreatic ductal system: a systematic review and metaanalysis of observational studies