Cancer Res Treat.
2023 Oct;55(4):1087-1095. 10.4143/crt.2023.682.
Clinicopathological Characteristics of NRG1 Fusion–Positive Solid Tumors in Korean Patients
- Affiliations
-
- 1Department of Pathology, Yonsei University College of Medicine, Seoul, Korea
- 2Department of Radiology, Yonsei University College of Medicine, Seoul, Korea
- 3Division of Medical Oncology, Department of Internal Medicine, Yonsei Cancer Center, Yonsei University College of Medicine, Seoul, Korea
- KMID: 2547783
- DOI: http://doi.org/10.4143/crt.2023.682
Abstract
- Purpose
Neuregulin 1 (NRG1) gene fusion is a potentially actionable oncogenic driver. The oncoprotein binds to ERBB3-ERBB2 heterodimers and activates downstream signaling, supporting a therapeutic approach for inhibiting ERBB3/ERBB2. However, the frequency and clinicopathological features of solid tumors harboring NRG1 fusions in Korean patients remain largely unknown.
Materials and Methods
We reviewed archival data from next-generation sequencing panel tests conducted at a single institution, specifically selecting patients with in-frame fusions that preserved the functional domain. The clinicopathological characteristics of patients harboring NRG1 fusions were retrospectively reviewed.
Results
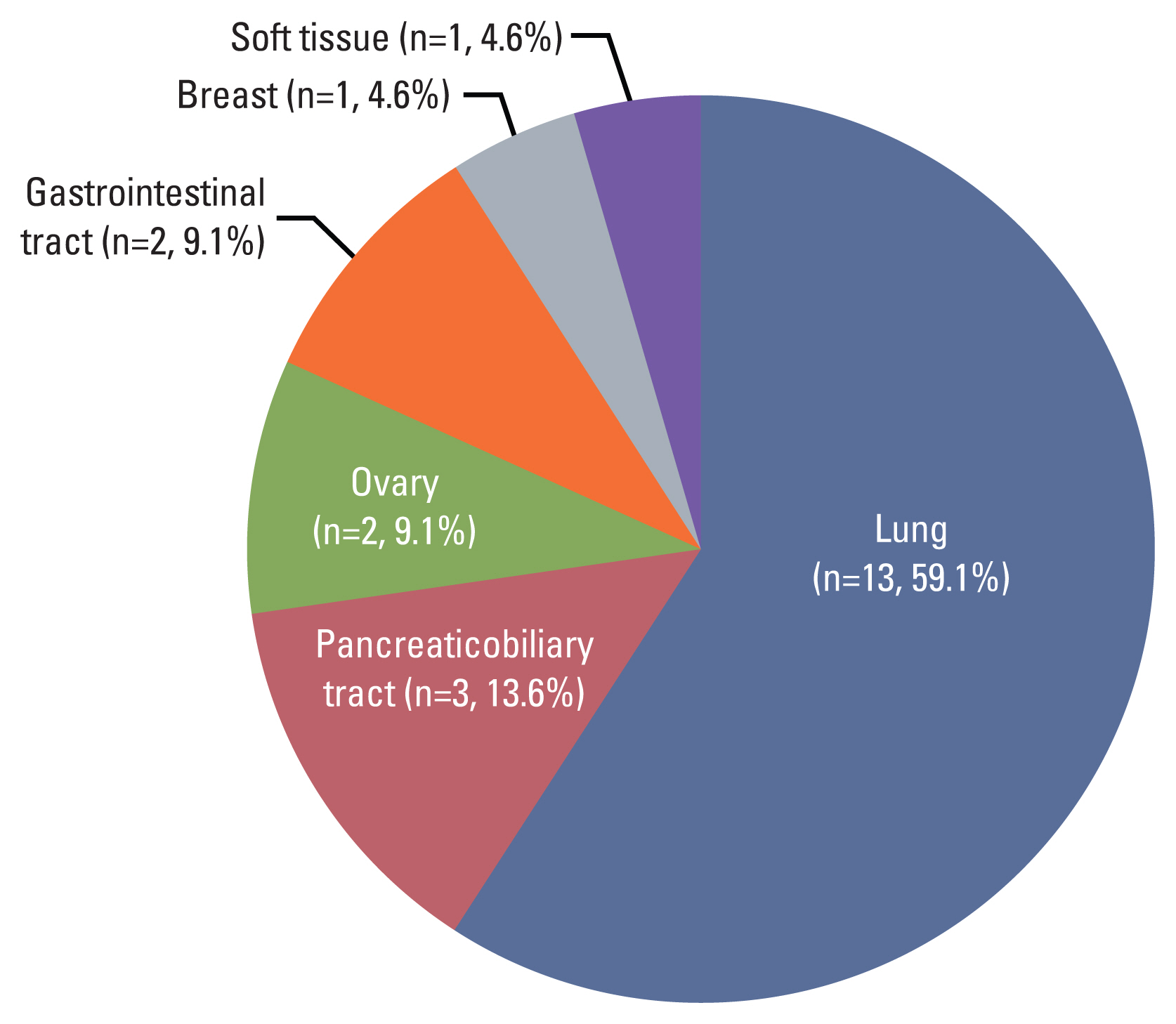
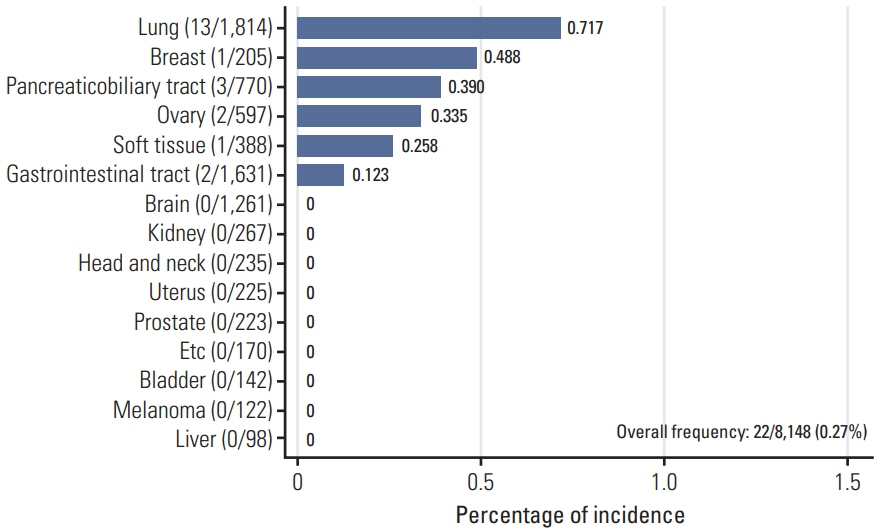
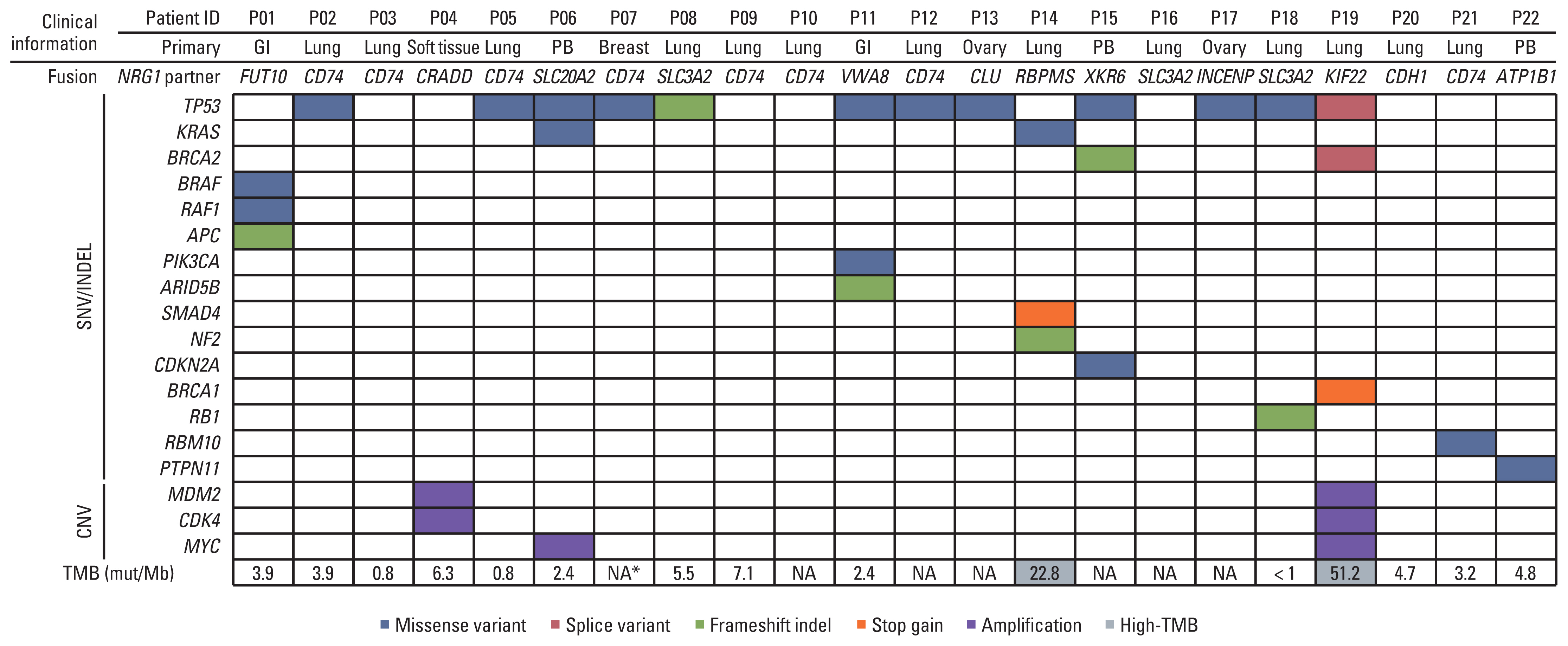
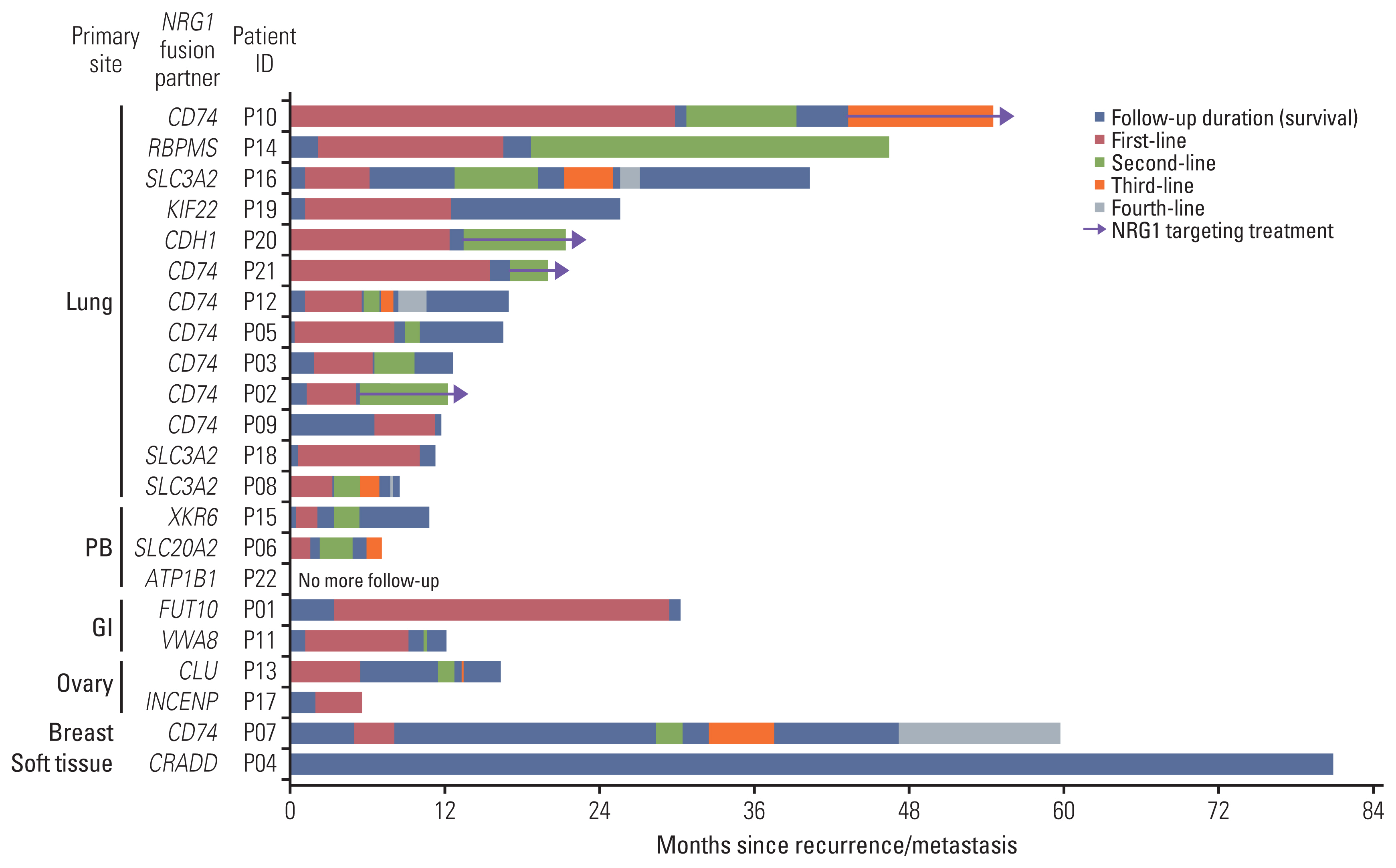
Out of 8,148 patients, NRG1 fusions were identified in 22 patients (0.27%). The average age of the patients was 59 years (range, 32 to 78 years), and the male-to-female ratio was 1:1.2. The lung was the most frequently observed primary site (n=13), followed by the pancreaticobiliary tract (n=3), gastrointestinal tract (n=2, stomach and rectum each), ovary (n=2), breast (n=1), and soft tissue (n=1). Histologically, all tumors demonstrated adenocarcinoma histology, with the exception of one case of sarcoma. CD74 (n=8) and SLC3A2 (n=4) were the most frequently identified fusion partners. Dominant features included the presence of fewer than three co-occurring genetic alterations, a low tumor mutation burden, and low programmed death-ligand 1 expression. Various clinical responses were observed in patients with NRG1 fusions.
Conclusion
Despite the rarity of NRG1 fusions in Korean patients with solid tumors, identification through next-generation sequencing enables the possibility of new targeted therapies.
Keyword
Figure
Reference
-
References
1. Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007; 7:233–45.2. Schram AM, Chang MT, Jonsson P, Drilon A. Fusions in solid tumours: diagnostic strategies, targeted therapy, and acquired resistance. Nat Rev Clin Oncol. 2017; 14:735–48.3. Tzahar E, Levkowitz G, Karunagaran D, Yi L, Peles E, Lavi S, et al. ErbB-3 and ErbB-4 function as the respective low and high affinity receptors of all Neu differentiation factor/heregulin isoforms. J Biol Chem. 1994; 269:25226–33.4. van Lengerich B, Agnew C, Puchner EM, Huang B, Jura N. EGF and NRG induce phosphorylation of HER3/ERBB3 by EGFR using distinct oligomeric mechanisms. Proc Natl Acad Sci U S A. 2017; 114:E2836–45.5. Liu X, Baker E, Eyre HJ, Sutherland GR, Zhou M. Gamma-heregulin: a fusion gene of DOC-4 and neuregulin-1 derived from a chromosome translocation. Oncogene. 1999; 18:7110–4.6. Schaefer G, Fitzpatrick VD, Sliwkowski MX. Gamma-heregulin: a novel heregulin isoform that is an autocrine growth factor for the human breast cancer cell line, MDA-MB-175. Oncogene. 1997; 15:1385–94.7. Huang HE, Chin SF, Ginestier C, Bardou VJ, Adelaide J, Iyer NG, et al. A recurrent chromosome breakpoint in breast cancer at the NRG1/neuregulin 1/heregulin gene. Cancer Res. 2004; 64:6840–4.8. Adelaide J, Huang HE, Murati A, Alsop AE, Orsetti B, Mozziconacci MJ, et al. A recurrent chromosome translocation breakpoint in breast and pancreatic cancer cell lines targets the neuregulin/NRG1 gene. Genes Chromosomes Cancer. 2003; 37:333–45.9. Fernandez-Cuesta L, Plenker D, Osada H, Sun R, Menon R, Leenders F, et al. CD74-NRG1 fusions in lung adenocarcinoma. Cancer Discov. 2014; 4:415–22.10. Shim HS, Kenudson M, Zheng Z, Liebers M, Cha YJ, Hoang Ho Q, et al. Unique genetic and survival characteristics of invasive mucinous adenocarcinoma of the lung. J Thorac Oncol. 2015; 10:1156–62.11. Drilon A, Duruisseaux M, Han JY, Ito M, Falcon C, Yang SR, et al. Clinicopathologic features and response to therapy of NRG1 fusion-driven lung cancers: the eNRGy1 global multicenter registry. J Clin Oncol. 2021; 39:2791–802.12. Jonna S, Feldman RA, Swensen J, Gatalica Z, Korn WM, Borghaei H, et al. Detection of NRG1 gene fusions in solid tumors. Clin Cancer Res. 2019; 25:4966–72.13. Cadranel J, Liu SV, Duruisseaux M, Branden E, Goto Y, Weinberg BA, et al. Therapeutic potential of afatinib in NRG1 fusion-driven solid tumors: a case series. Oncologist. 2021; 26:7–16.14. Fontana E, Torga G, Fostea R, Cleator S, Wasserman E, Murat A, et al. Sustained tumor regression with zenocutuzumab, a bispecific antibody targeting human epidermal growth factor receptor 2/human epidermal growth factor receptor 3 signaling, in NRG1 fusion-positive, estrogen receptor-positive breast cancer after progression on a cyclin-dependent kinase 4/6 inhibitor. JCO Precis Oncol. 2022; 6:e2100446.15. Schram AM, Odintsov I, Espinosa-Cotton M, Khodos I, Sisso WJ, Mattar MS, et al. Zenocutuzumab, a HER2xHER3 bispecific antibody, is effective therapy for tumors driven by NRG1 gene rearrangements. Cancer Discov. 2022; 12:1233–47.16. Yatabe Y, Mitsudomi T, Takahashi T. TTF-1 expression in pulmonary adenocarcinomas. Am J Surg Pathol. 2002; 26:767–73.17. Matsubara D, Soda M, Yoshimoto T, Amano Y, Sakuma Y, Yamato A, et al. Inactivating mutations and hypermethylation of the NKX2-1/TTF-1 gene in non-terminal respiratory unit-type lung adenocarcinomas. Cancer Sci. 2017; 108:1888–96.18. Park WY, Kim MH, Shin DH, Lee JH, Choi KU, Kim JY, et al. Ciliated adenocarcinomas of the lung: a tumor of non-terminal respiratory unit origin. Mod Pathol. 2012; 25:1265–74.19. Park E, Shim HS. Detection of targetable genetic alterations in Korean lung cancer patients: a comparison study of single-gene assays and targeted next-generation sequencing. Cancer Res Treat. 2020; 52:543–51.20. Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017; 19:4–23.21. Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Kallberg M, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016; 32:1220–2.22. Uhrig S, Ellermann J, Walther T, Burkhardt P, Frohlich M, Hutter B, et al. Accurate and efficient detection of gene fusions from RNA sequencing data. Genome Res. 2021; 31:448–60.23. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21.24. Wen D, Suggs SV, Karunagaran D, Liu N, Cupples RL, Luo Y, et al. Structural and functional aspects of the multiplicity of Neu differentiation factors. Mol Cell Biol. 1994; 14:1909–19.25. Geles A, Gruber-Moesenbacher U, Quehenberger F, Manzl C, Al Effah M, Grygar E, et al. Pulmonary mucinous adenocarcinomas: architectural patterns in correlation with genetic changes, prognosis and survival. Virchows Arch. 2015; 467:675–86.26. Snyder EL, Watanabe H, Magendantz M, Hoersch S, Chen TA, Wang DG, et al. Nkx2-1 represses a latent gastric differentiation program in lung adenocarcinoma. Mol Cell. 2013; 50:185–99.27. Heining C, Horak P, Uhrig S, Codo PL, Klink B, Hutter B, et al. NRG1 fusions in KRAS wild-type pancreatic cancer. Cancer Discov. 2018; 8:1087–95.28. Shin DH, Kim SH, Choi M, Bae YK, Han C, Choi BK, et al. Oncogenic KRAS promotes growth of lung cancer cells expressing SLC3A2-NRG1 fusion via ADAM17-mediated shedding of NRG1. Oncogene. 2022; 41:280–92.29. Geuijen CA, De Nardis C, Maussang D, Rovers E, Gallenne T, Hendriks LJA, et al. Unbiased combinatorial screening identifies a bispecific IgG1 that potently inhibits HER3 signaling via HER2-guided ligand blockade. Cancer Cell. 2018; 33:922–36.30. Schram AM, Goto K, Kim DW, Martin-Romano P, Ou SH, O’Kane GM, et al. Efficacy and safety of zenocutuzumab, a HER2 × HER3 bispecific antibody, across advanced NRG1 fusion (NRG1+) cancers. J Clin Oncol. 2022; 40(16 Suppl):105.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Unpredictable Spontaneous Fusion after Percutaneous Vertebroplasty and Kyphoplasty in Osteoporotic Compression Fracture
- Usefulness of Chimeric Transcript in the Diagnosis of Pediatric Solid Tumors
- Exploring the clinicopathological parameters of HER2 low breast cancers: insights from a retrospective cohort study
- Lymphodepletion in Chimeric Antigen Receptor T-Cell Therapy for Solid Tumors: A Focus on Brain Tumors
- Neuregulin 1/ErbB4 signaling attenuates neuronal cell damage under oxygen-glucose deprivation in primary hippocampal neurons