Endocrinol Metab.
2023 Aug;38(4):445-454. 10.3803/EnM.2023.1702.
Different Molecular Phenotypes of Progression in BRAF- and RAS-Like Papillary Thyroid Carcinoma
- Affiliations
-
- 1Department of Medicine, CHA University School of Medicine, Seongnam, Korea
- 2Department of Biomedical Science, Graduate School, CHA University, Seongnam, Korea
- 3Department of Internal Medicine, Seoul Metropolitan Government Seoul National University Boramae Medical Center, Seoul, Korea
- 4Department of Internal Medicine, CHA Bundang Medical Center, CHA University School of Medicine, Seongnam, Korea
- KMID: 2545277
- DOI: http://doi.org/10.3803/EnM.2023.1702
Abstract
- Background
Papillary thyroid carcinoma (PTC) can be classified into two distinct molecular subtypes, BRAF-like (BL) and RASlike (RL). However, the molecular characteristics of each subtype according to clinicopathological factors have not yet been determined. We aimed to investigate the gene signatures and tumor microenvironment according to clinicopathological factors, and to identify the mechanism of progression in BL-PTCs and RL-PTCs.
Methods
We analyzed RNA sequencing data and corresponding clinicopathological information of 503 patients with PTC from The Cancer Genome Atlas database. We performed differentially expressed gene (DEG), Gene Ontology, and molecular pathway enrichment analyses according to clinicopathological factors in each molecular subtype. EcoTyper and CIBERSORTx were used to deconvolve the tumor cell types and their surrounding microenvironment.
Results
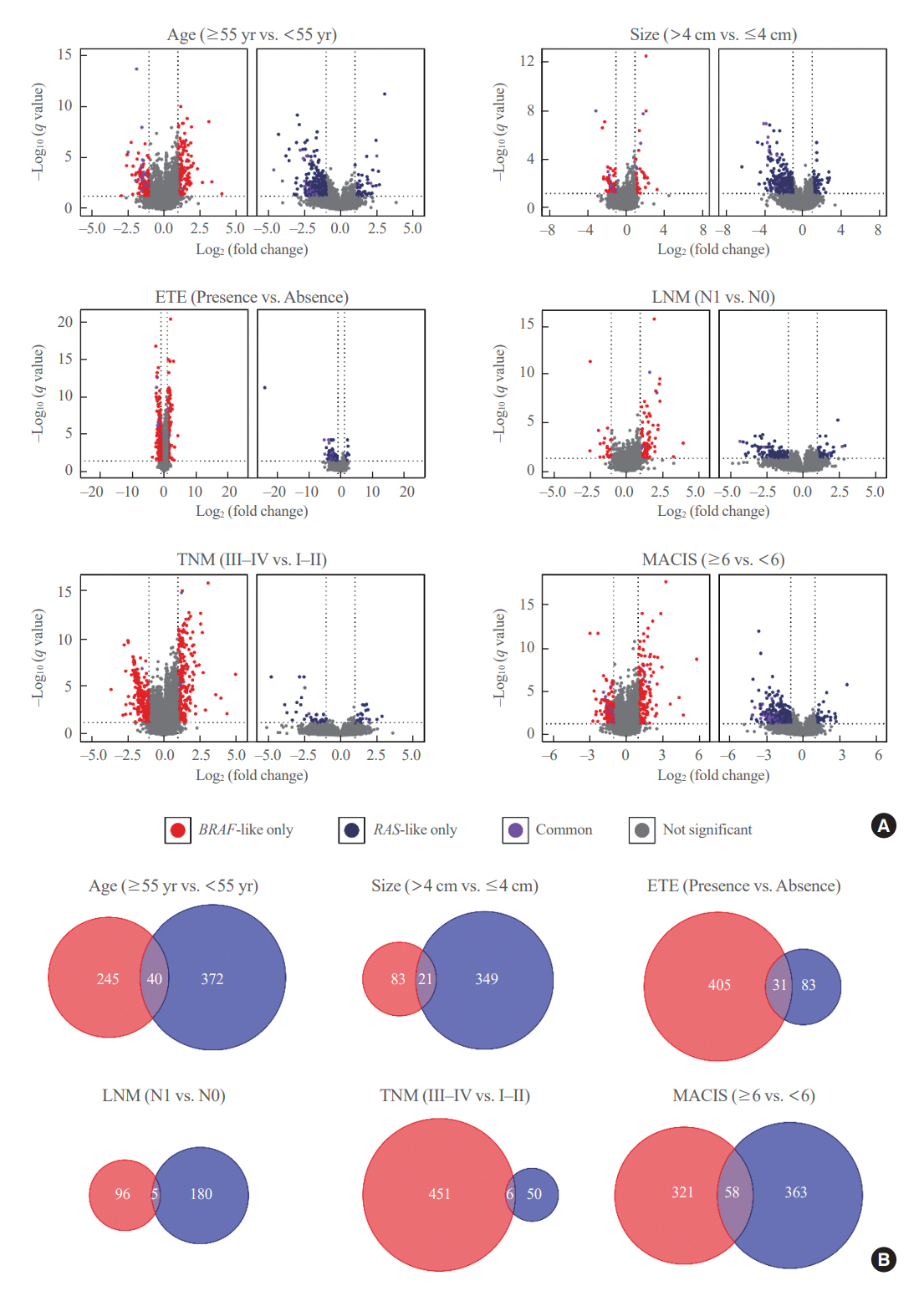
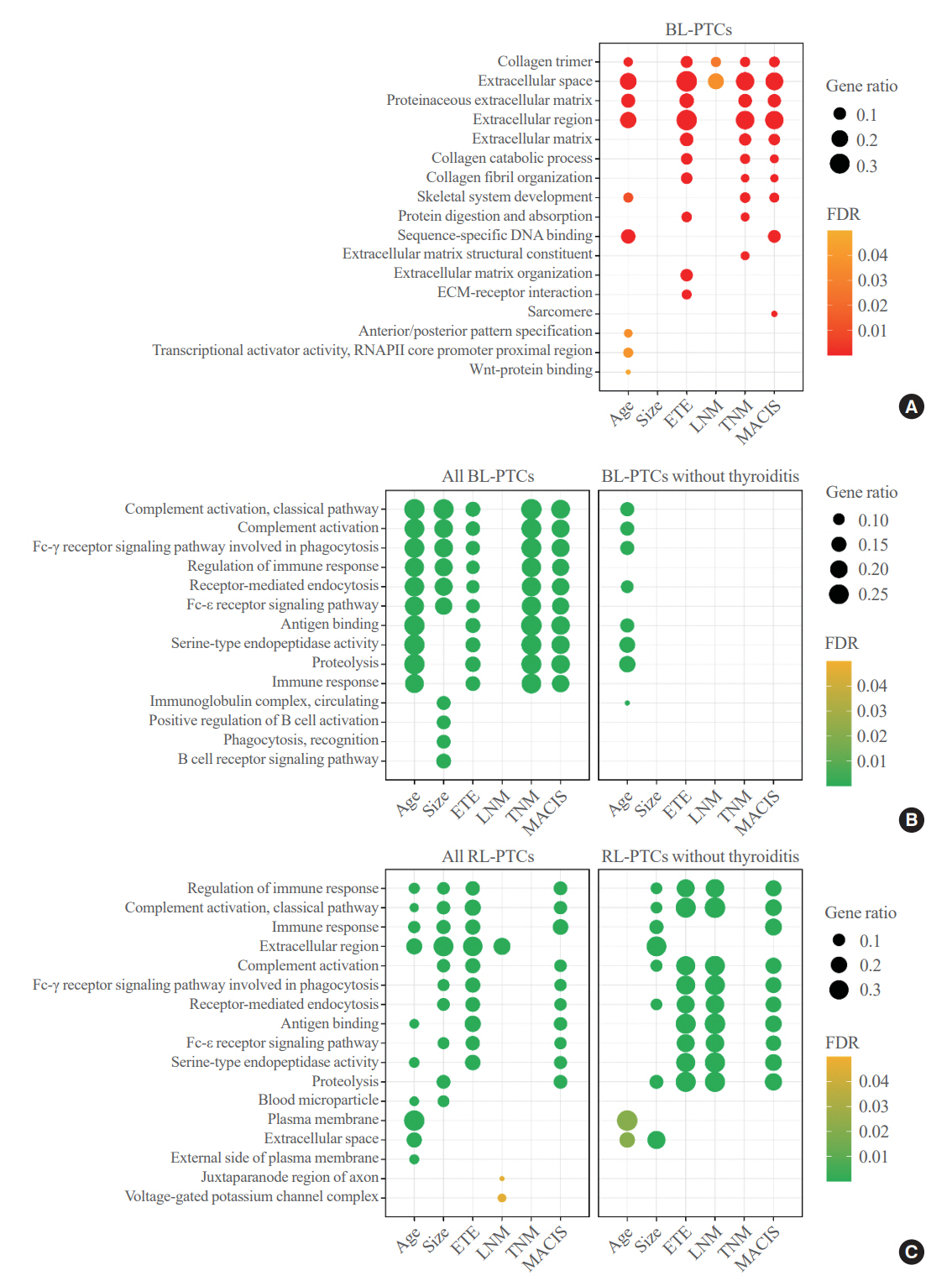
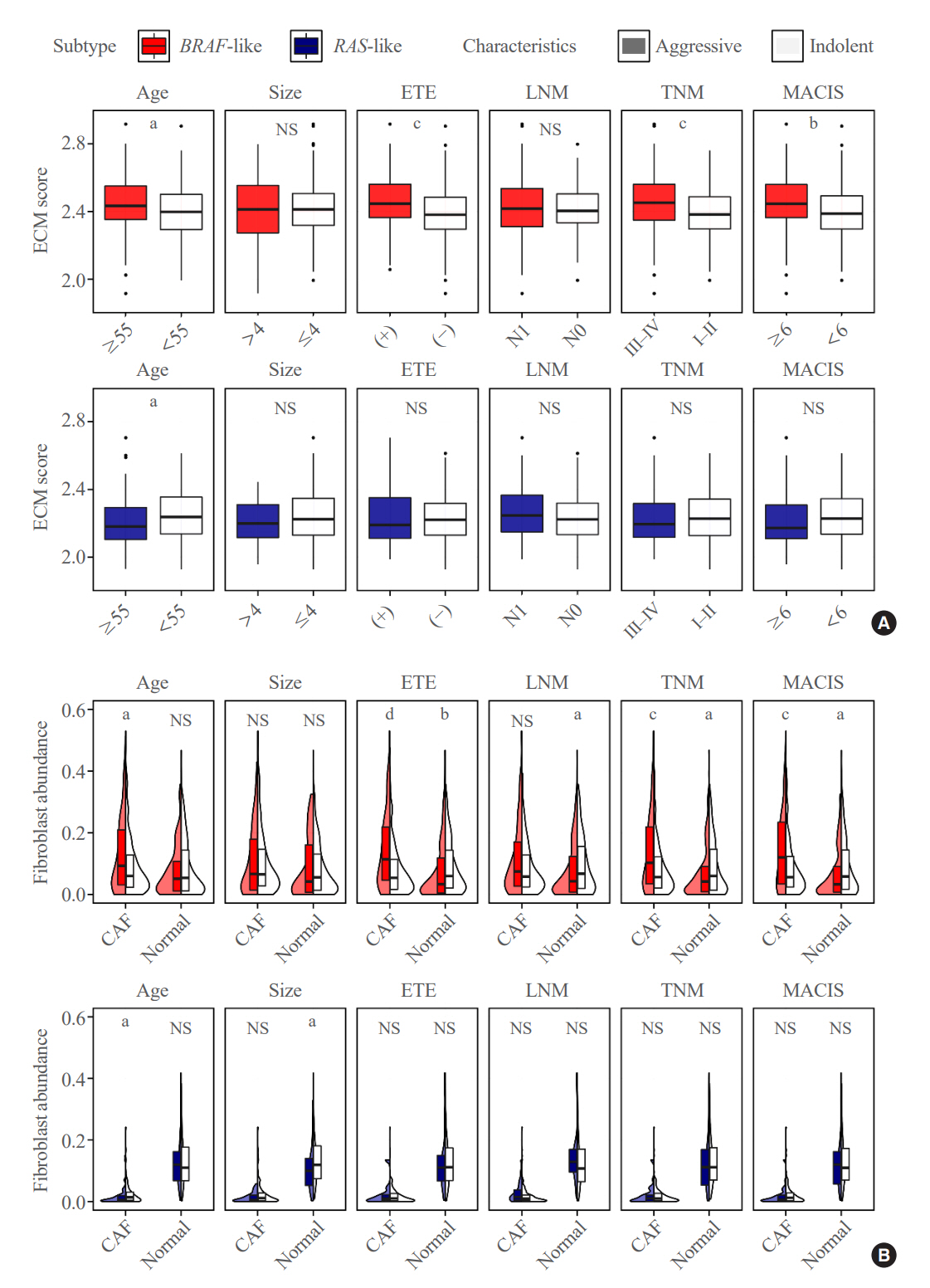
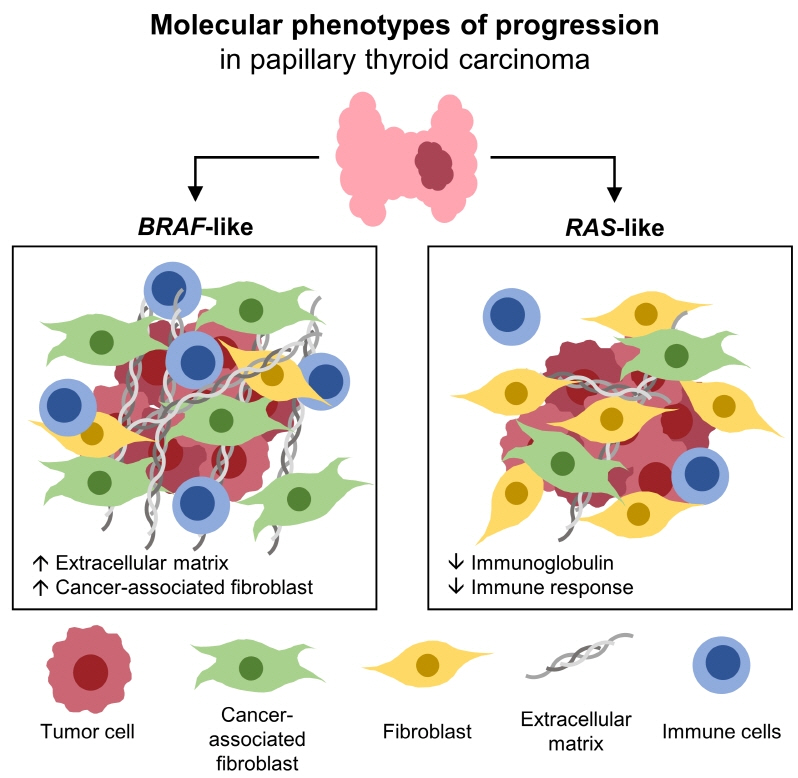
Even for the same clinicopathological factors, overlapping DEGs between the two molecular subtypes were uncommon, indicating that BL-PTCs and RL-PTCs have different progression mechanisms. Genes related to the extracellular matrix were commonly upregulated in BL-PTCs with aggressive clinicopathological factors, such as old age (≥55 years), presence of extrathyroidal extension, lymph node metastasis, advanced tumor-node-metastasis (TNM) stage, and high metastasis-age-completeness of resection- invasion-size (MACIS) scores (≥6). Furthermore, in the deconvolution analysis of tumor microenvironment, cancer-associated fibroblasts were significantly enriched. In contrast, in RL-PTCs, downregulation of immune response and immunoglobulin-related genes was significantly associated with aggressive characteristics, even after adjusting for thyroiditis status.
Conclusion
The molecular phenotypes of cancer progression differed between BL-PTC and RL-PTC. In particular, extracellular matrix and cancer-associated fibroblasts, which constitute the tumor microenvironment, would play an important role in the progression of BL-PTC that accounts for the majority of advanced PTCs.
Figure
Reference
-
1. Davies L, Welch HG. Current thyroid cancer trends in the United States. JAMA Otolaryngol Head Neck Surg. 2014; 140:317–22.2. Amin MB, Edge SB, Greene FL, Bryd DR, Brookland RK, Washington MK, et al. AJCC cancer staging manual. 8th ed. New York: Springer;2017.3. Grogan RH, Kaplan SP, Cao H, Weiss RE, Degroot LJ, Simon CA, et al. A study of recurrence and death from papillary thyroid cancer with 27 years of median follow-up. Surgery. 2013; 154:1436–47.4. Nixon IJ, Wang LY, Migliacci JC, Eskander A, Campbell MJ, Aniss A, et al. An international multi-institutional validation of age 55 years as a cutoff for risk stratification in the AJCC/UICC staging system for well-differentiated thyroid cancer. Thyroid. 2016; 26:373–80.5. Leboulleux S, Rubino C, Baudin E, Caillou B, Hartl DM, Bidart JM, et al. Prognostic factors for persistent or recurrent disease of papillary thyroid carcinoma with neck lymph node metastases and/or tumor extension beyond the thyroid capsule at initial diagnosis. J Clin Endocrinol Metab. 2005; 90:5723–9.6. Hay ID, Bergstralh EJ, Goellner JR, Ebersold JR, Grant CS. Predicting outcome in papillary thyroid carcinoma: development of a reliable prognostic scoring system in a cohort of 1779 patients surgically treated at one institution during 1940 through 1989. Surgery. 1993; 114:1050–8.7. Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014; 159:676–90.8. Yoo SK, Lee S, Kim SJ, Jee HG, Kim BA, Cho H, et al. Comprehensive analysis of the transcriptional and mutational landscape of follicular and papillary thyroid cancers. PLoS Genet. 2016; 12:e1006239.9. Song YS, Won JK, Yoo SK, Jung KC, Kim MJ, Kim SJ, et al. Comprehensive transcriptomic and genomic profiling of subtypes of follicular variant of papillary thyroid carcinoma. Thyroid. 2018; 28:1468–78.10. Song YS, Park YJ. Genomic characterization of differentiated thyroid carcinoma. Endocrinol Metab (Seoul). 2019; 34:1–10.11. Whiteside TL. The tumor microenvironment and its role in promoting tumor growth. Oncogene. 2008; 27:5904–12.12. Jolly LA, Novitskiy S, Owens P, Massoll N, Cheng N, Fang W, et al. Fibroblast-mediated collagen remodeling within the tumor microenvironment facilitates progression of thyroid cancers driven by BrafV600E and Pten loss. Cancer Res. 2016; 76:1804–13.13. Minna E, Brich S, Todoerti K, Pilotti S, Collini P, Bonaldi E, et al. Cancer associated fibroblasts and senescent thyroid cells in the invasive front of thyroid carcinoma. Cancers (Basel). 2020; 12:112.14. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012; 2:401–4.15. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550.16. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009; 4:44–57.17. Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009; 462:108–12.18. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005; 102:15545–50.19. Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015; 12:453–7.20. Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019; 37:773–82.21. Luca BA, Steen CB, Matusiak M, Azizi A, Varma S, Zhu C, et al. Atlas of clinically distinct cell states and ecosystems across human solid tumors. Cell. 2021; 184:5482–96.22. Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017; 18:220.23. Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013; 4:2612.24. Becht E, Giraldo NA, Lacroix L, Buttard B, Elarouci N, Petitprez F, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016; 17:218.25. Racle J, Gfeller D. EPIC: a tool to estimate the proportions of different cell types from bulk gene expression data. Methods Mol Biol. 2020; 2120:233–48.26. Sahai E, Astsaturov I, Cukierman E, DeNardo DG, Egeblad M, Evans RM, et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer. 2020; 20:174–86.27. Pu W, Shi X, Yu P, Zhang M, Liu Z, Tan L, et al. Single-cell transcriptomic analysis of the tumor ecosystems underlying initiation and progression of papillary thyroid carcinoma. Nat Commun. 2021; 12:6058.28. Bergdorf K, Ferguson DC, Mehrad M, Ely K, Stricker T, Weiss VL. Papillary thyroid carcinoma behavior: clues in the tumor microenvironment. Endocr Relat Cancer. 2019; 26:601–14.29. Sarvaria A, Madrigal JA, Saudemont A. B cell regulation in cancer and anti-tumor immunity. Cell Mol Immunol. 2017; 14:662–74.30. Mauri C, Bosma A. Immune regulatory function of B cells. Annu Rev Immunol. 2012; 30:221–41.31. Hsu HM, Chu CM, Chang YJ, Yu JC, Chen CT, Jian CE, et al. Six novel immunoglobulin genes as biomarkers for better prognosis in triple-negative breast cancer by gene co-expression network analysis. Sci Rep. 2019; 9:4484.32. Knauf JA, Sartor MA, Medvedovic M, Lundsmith E, Ryder M, Salzano M, et al. Progression of BRAF-induced thyroid cancer is associated with epithelial-mesenchymal transition requiring concomitant MAP kinase and TGFb signaling. Oncogene. 2011; 30:3153–62.33. Dong C, Davis RJ, Flavell RA. MAP kinases in the immune response. Annu Rev Immunol. 2002; 20:55–72.34. Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC, Abraham RT. The PI3K pathway in human disease. Cell. 2017; 170:605–35.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Genetic Alterations in Follicular Cell-derived Thyroid Carcinomas
- Diagnostic Dilemma of a Follicular Lesions/Neoplasm in Thyroid Fine Needle Aspiration Cytology
- A Case of Multifocal Papillary Thyroid Carcinoma Consisting of One Encapsulated Follicular Variant with BRAF K601E Mutation and Three Conventional Types with BRAF V600E Mutation
- Medullary and Papillary Thyroid Carcinoma as a Collision Tumor: Report of Five Cases
- ras Mutation in Korean Papillary Thyroid Carcinomas