Healthc Inform Res.
2023 Jul;29(3):246-255. 10.4258/hir.2023.29.3.246.
Development and Verification of Time-Series Deep Learning for Drug-Induced Liver Injury Detection in Patients Taking Angiotensin II Receptor Blockers: A Multicenter Distributed Research Network Approach
- Affiliations
-
- 1Department of Biomedical Systems Informatics, Yonsei University College of Medicine, Seoul, Korea
- 2Medical Informatics Collaborative Unit, Department of Research Affairs, Yonsei University College of Medicine, Seoul, Korea
- 3Healthcare Data Science Center, Konyang University Hospital, Daejeon, Korea
- 4Department of Biomedical Sciences, Ajou University Graduate School of Medicine, Seoul, Korea
- 5Department of Statistics, Korea University, Suwon, Korea
- 6Healthcare AI Team, National Cancer Center, Goyang, Korea
- 7Transdisciplinary Department of Medicine & Advanced Technology, Seoul National University Hospital, Seoul, Korea
- 8Division of Allergy and Immunology, Department of Internal Medicine, Institute of Allergy, Yonsei University College of Medicine, Seoul, Korea
- KMID: 2544876
- DOI: http://doi.org/10.4258/hir.2023.29.3.246
Abstract
Objectives
The objective of this study was to develop and validate a multicenter-based, multi-model, time-series deep learning model for predicting drug-induced liver injury (DILI) in patients taking angiotensin receptor blockers (ARBs). The study leveraged a national-level multicenter approach, utilizing electronic health records (EHRs) from six hospitals in Korea.
Methods
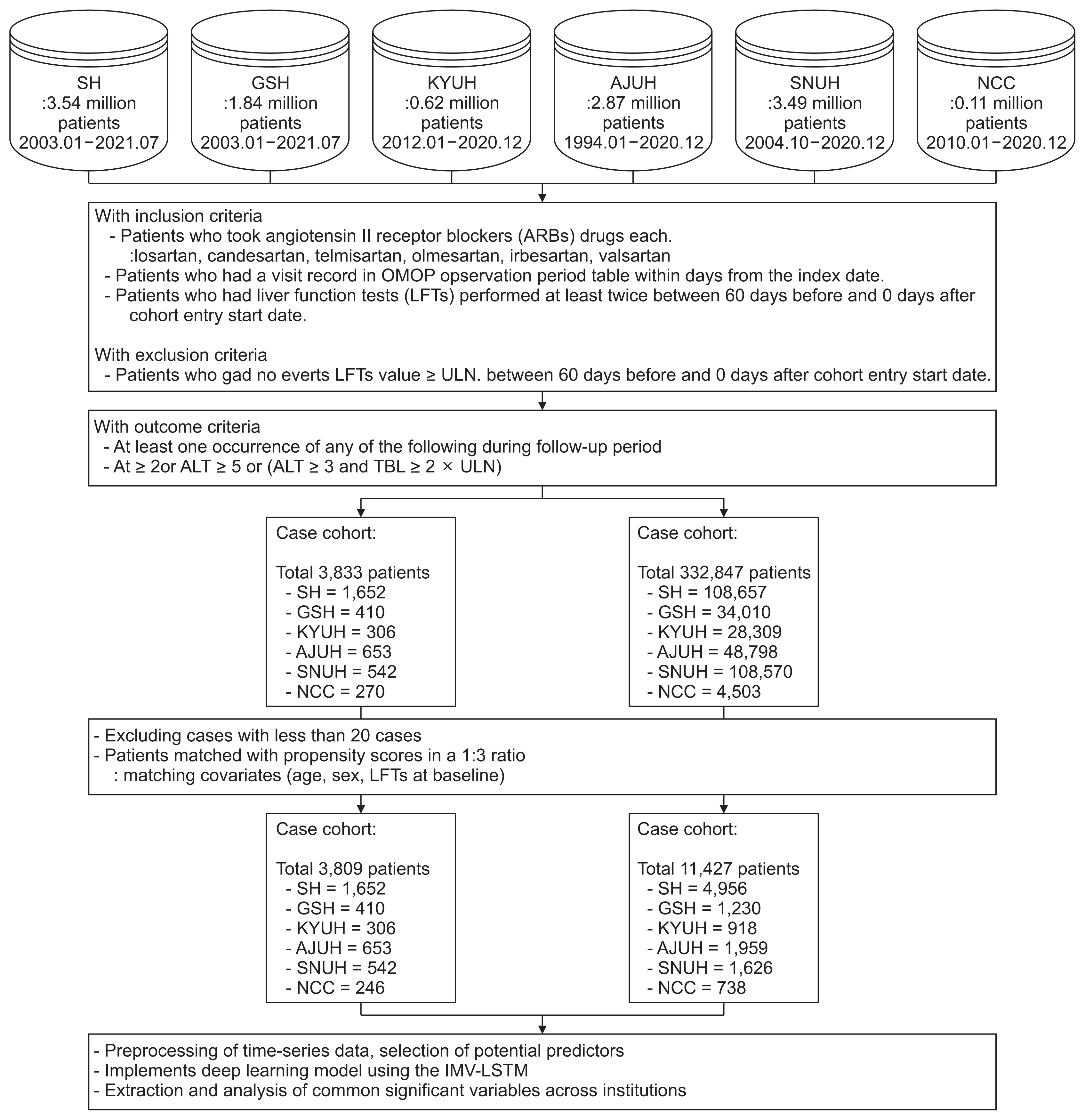
A retrospective cohort analysis was conducted using EHRs from six hospitals in Korea, comprising a total of 10,852 patients whose data were converted to the Common Data Model. The study assessed the incidence rate of DILI among patients taking ARBs and compared it to a control group. Temporal patterns of important variables were analyzed using an interpretable timeseries model.
Results
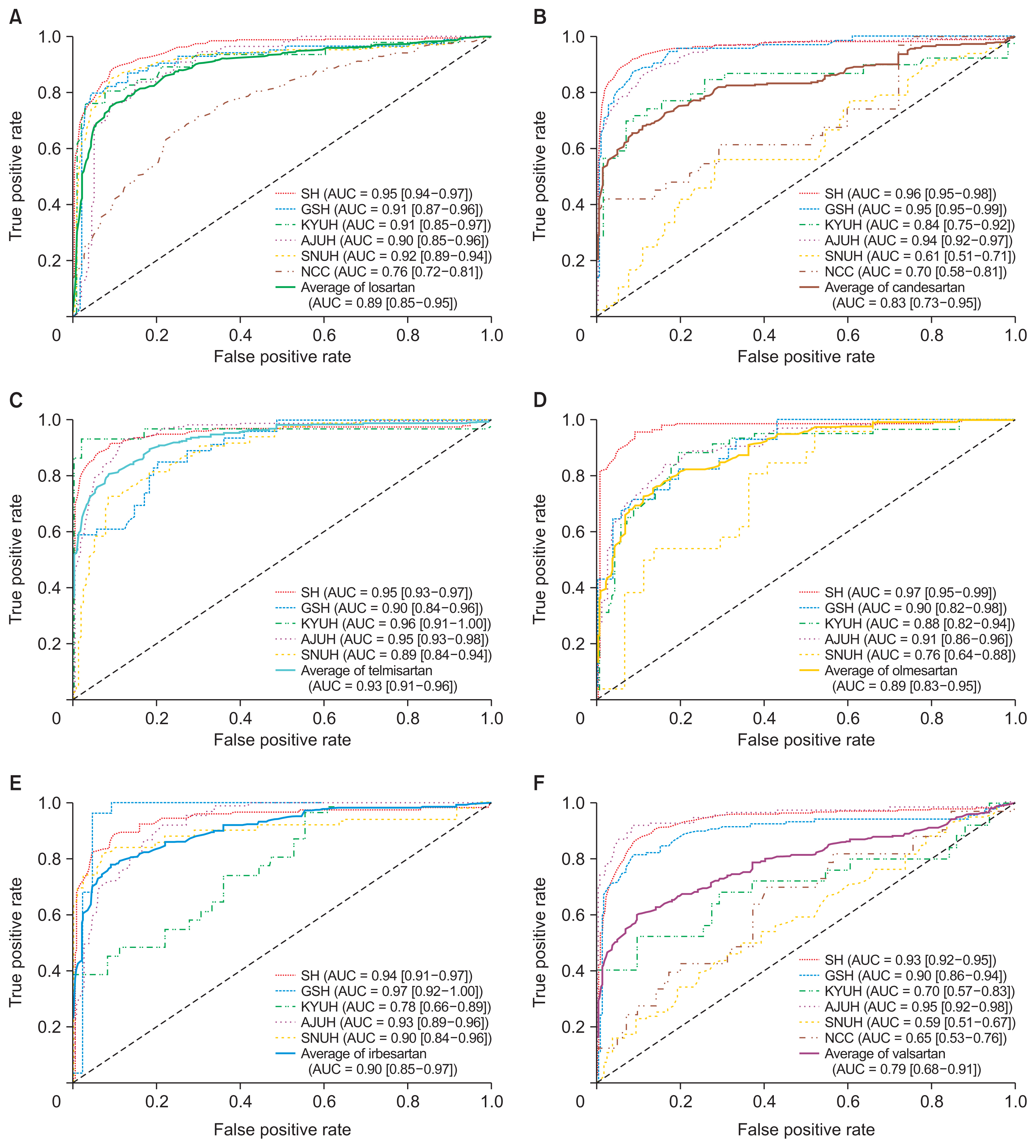
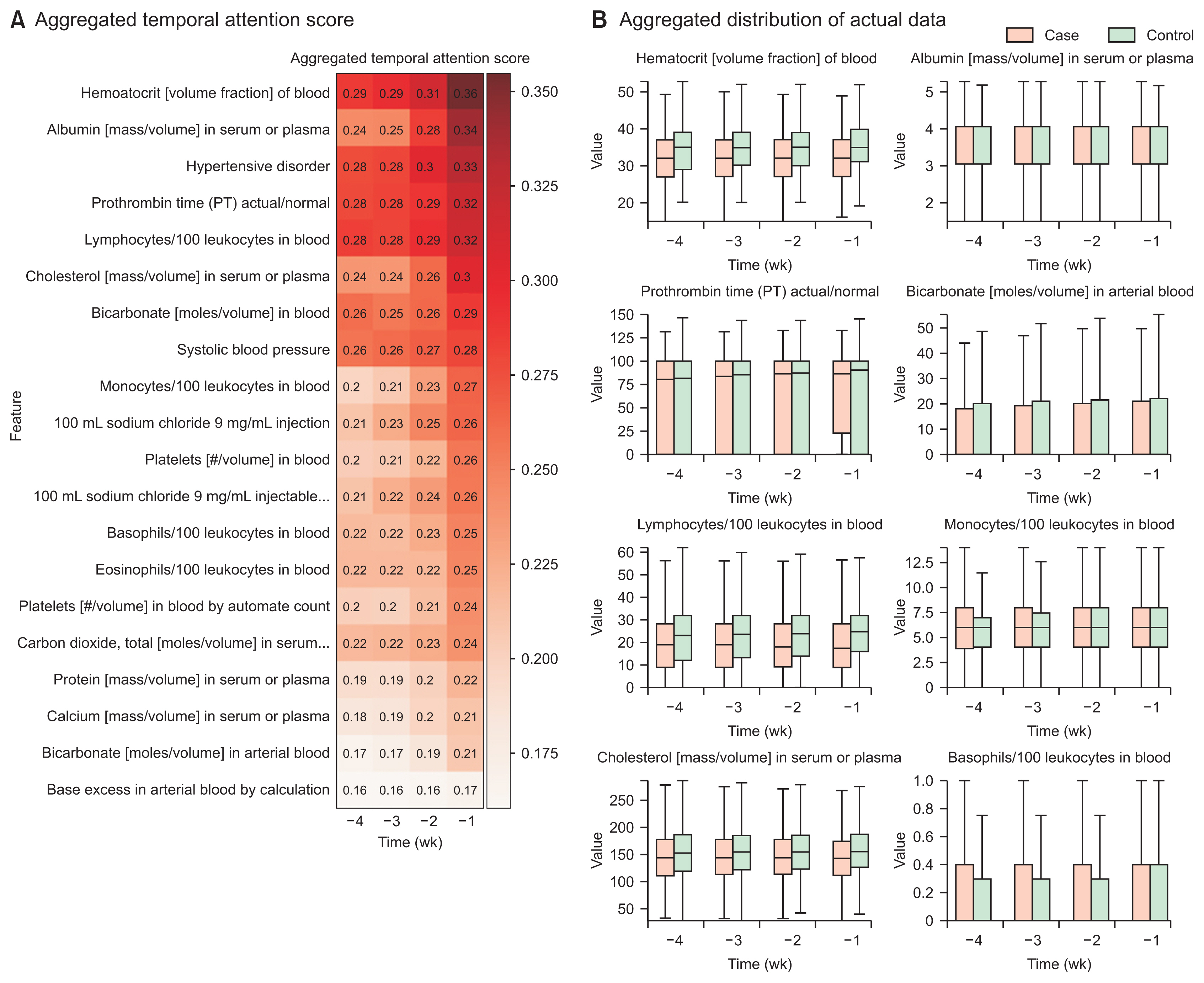
The overall incidence rate of DILI among patients taking ARBs was found to be 1.09%. The incidence rates varied for each specific ARB drug and institution, with valsartan having the highest rate (1.24%) and olmesartan having the lowest rate (0.83%). The DILI prediction models showed varying performance, measured by the average area under the receiver operating characteristic curve, with telmisartan (0.93), losartan (0.92), and irbesartan (0.90) exhibiting higher classification performance. The aggregated attention scores from the models highlighted the importance of variables such as hematocrit, albumin, prothrombin time, and lymphocytes in predicting DILI.
Conclusions
Implementing a multicenter-based timeseries classification model provided evidence that could be valuable to clinicians regarding temporal patterns associated with DILI in ARB users. This information supports informed decisions regarding appropriate drug use and treatment strategies.
Figure
Reference
-
References
1. Liu Y, Aickelin U. Feature selection in detection of adverse drug reactions from the Health Improvement Network (THIN) database. arXiv [Preprint]. 2014. Sep. 2. https://doi.org/10.48550/arXiv.1409.0775.
Article2. Montastruc JL, Lafaurie M, de Canecaude C, Durrieu G, Sommet A, Montastruc F, et al. Fatal adverse drug reactions: a worldwide perspective in the World Health Organization pharmacovigilance database. Br J Clin Pharmacol. 2021; 87(11):4334–40. https://doi.org/10.1111/bcp.14851.
Article3. Sonawane KB, Cheng N, Hansen RA. Serious adverse drug events reported to the FDA: analysis of the FDA adverse event reporting system 2006–2014 database. J Manag Care Spec Pharm. 2018; 24(7):682–90. https://doi.org/10.18553/jmcp.2018.24.7.682.
Article4. Jaganathan K, Tayara H, Chong KT. Prediction of drug-induced liver toxicity using SVM and optimal descriptor sets. Int J Mol Sci. 2021; 22(15):8073. https://doi.org/10.3390/ijms22158073.
Article5. Liu A, Walter M, Wright P, Bartosik A, Dolciami D, Elbasir A, et al. Prediction and mechanistic analysis of drug-induced liver injury (DILI) based on chemical structure. Biol Direct. 2021; 16(1):6. https://doi.org/10.1186/s13062-020-00285-0.
Article6. Chalasani N, Bjornsson E. Risk factors for idiosyncratic drug-induced liver injury. Gastroenterology. 2010; 138(7):2246–59. https://doi.org/10.1053/j.gastro.2010.04.001.
Article7. Chen Z, Jiang Y, Zhang X, Zheng R, Qiu R, Sun Y, et al. The prediction approach of drug-induced liver injury: response to the issues of reproducible science of artificial intelligence in real-world applications. Brief Bioinform. 2022. 23(4):bbac196. https://doi.org/10.1093/bib/bbac196.
Article8. Fleurence RL, Curtis LH, Califf RM, Platt R, Selby JV, Brown JS. Launching PCORnet, a national patient-centered clinical research network. J Am Med Inform Assoc. 2014; 21(4):578–82. https://doi.org/10.1136/amiajnl-2014-002747.
Article9. Huser V, Amos L. Analyzing real-world use of research common data elements. AMIA Annu Symp Proc. 2018; 2018:602–8.10. Voss EA, Makadia R, Matcho A, Ma Q, Knoll C, Schuemie M, et al. Feasibility and utility of applications of the common data model to multiple, disparate observational health databases. J Am Med Inform Assoc. 2015; 22(3):553–64. https://doi.org/10.1093/jamia/ocu023.
Article11. Ryu B, Yoo S, Kim S, Choi J. Development of prediction models for unplanned hospital readmission within 30 days based on common data model: a feasibility study. Methods Inf Med. 2021; 60(S 02):e65–75. https://doi.org/10.1055/s-0041-1735166.
Article12. Ball R, Robb M, Anderson SA, Dal Pan G. The FDA’s sentinel initiative: a comprehensive approach to medical product surveillance. Clin Pharmacol Ther. 2016; 99(3):265–8. https://doi.org/10.1002/cpt.320.
Article13. The Council for International Organizations of Medical Sciences (CIOMS). Drug-induced liver injury (DILI): current status and future directions for drug development and the post-market setting. Geneva, Switzerland: CIOMS;2020. https://doi.org/10.56759/ojsg8296.14. Goldstein A, Kapelner A, Bleich J, Pitkin E. Peeking inside the black box: Visualizing statistical learning with plots of individual conditional expectation. J Comput Graph Stat. 2015; 24(1):44–65. https://doi.org/10.1080/10618600.2014.907095.
Article15. Lundberg SM, Lee SI. A unified approach to interpreting model predictions. Adv Neural Inf Process Syst. 2017; 30:4765–74.16. Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-CAM: visual explanations from deep networks via gradient-based localization. In : Proceedings of the IEEE International Conference on Computer Vision; 2017 Oct 22–29; Venice, Italy. p. 618–26. https://doi.org/10.1109/ICCV.2017.74.
Article17. Guo T, Lin T, Antulov-Fantulin N. Exploring interpretable LSTM neural networks over multi-variable data. In : Proceedings of the 36th International Conference on Machine Learning (ICML); 2019 Jun 9–15; Long Beach, CA. p. 2494–504.18. Barreras A, Gurk-Turner C. Angiotensin II receptor blockers. Proc (Bayl Univ Med Cent). 2003; 16(1):123–6. https://doi.org/10.1080/08998280.2003.11927893.
Article19. Hill RD, Vaidya PN. Angiotensin II receptor blockers (ARB). Treasure Island (FL): StatPearls Publishing;2019.20. DigitalHealthcareLab. MOACDM [Internet]. Seoul, Korea: DigitalHealthcareLab;2023. [cited at 2023 Jul 27]. Available from: https://github.com/DigitalHealthcareLab/22MOACDM.21. Wang SV, Gagne JJ, Maro JC, Eworuke E, Kattinakere S, Kulldorff M, et al. Development and evaluation of a global propensity score for data mining with tree-based scan statistics (Sentinel Methods Protocol). Toledo (OH): The Sentinel System;2018.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Corrigendum to: Development and Verification of Time-Series Deep Learning for Drug-Induced Liver Injury Detection in Patients Taking Angiotensin II Receptor Blockers: A Multicenter Distributed Research Network Approach
- Are Angiotensin II Receptor Blockers Really Safe From Aminotransferase Elevation or Drug-Induced Liver Injury?
- Hyperkalemia from preoperative non-steroidal anti-inflammatory drugs and angiotensin II receptor blockers in patients with nephropathy
- Effects of Angiotensin II on ZO-1 in Glomerular Epithelial Cells
- Acute Renal Failure with Pulmonary Edema Induced by the Treatment of Angiotensin-Converting Enzyme Inhibitor and Angiotensin II Receptor Blocker in a Patient with Congenital Solitary Kidney