J Pathol Transl Med.
2023 May;57(3):147-157. 10.4132/jptm.2022.02.19.
Expression of specific microRNAs in tissue and plasma in colorectal cancer
- Affiliations
-
- 1The Graduate School, University of Santo Tomas, Manila, Philippines
- 2Department of Pathology and Laboratories, Mariano Marcos Memorial Hospital and Medical Center, Batac, Philippines
- 3Department of Surgery, Mariano Marcos Memorial Hospital and Medical Center, Batac, Philippines
- 4Research Center for Natural and Applied Sciences, University of Santo Tomas, Manila, Philippines
- 5Department of Biological Sciences, College of Science, University of Santo Tomas, Manila, Philippines
- KMID: 2542281
- DOI: http://doi.org/10.4132/jptm.2022.02.19
Abstract
- Background
MicroRNAs (miRNA/miR) play significant roles in the regulation of cell differentiation, cell cycle progression, and apoptosis. They become dysregulated during carcinogenesis and are eventually released into the circulation, enabling their detection in body fluids. Thus, this study compared the miRNA expression in tissue and plasma samples of colorectal cancer (CRC) patients and clinically healthy controls and determined miRNA expression as a potential CRC biomarker.
Methods
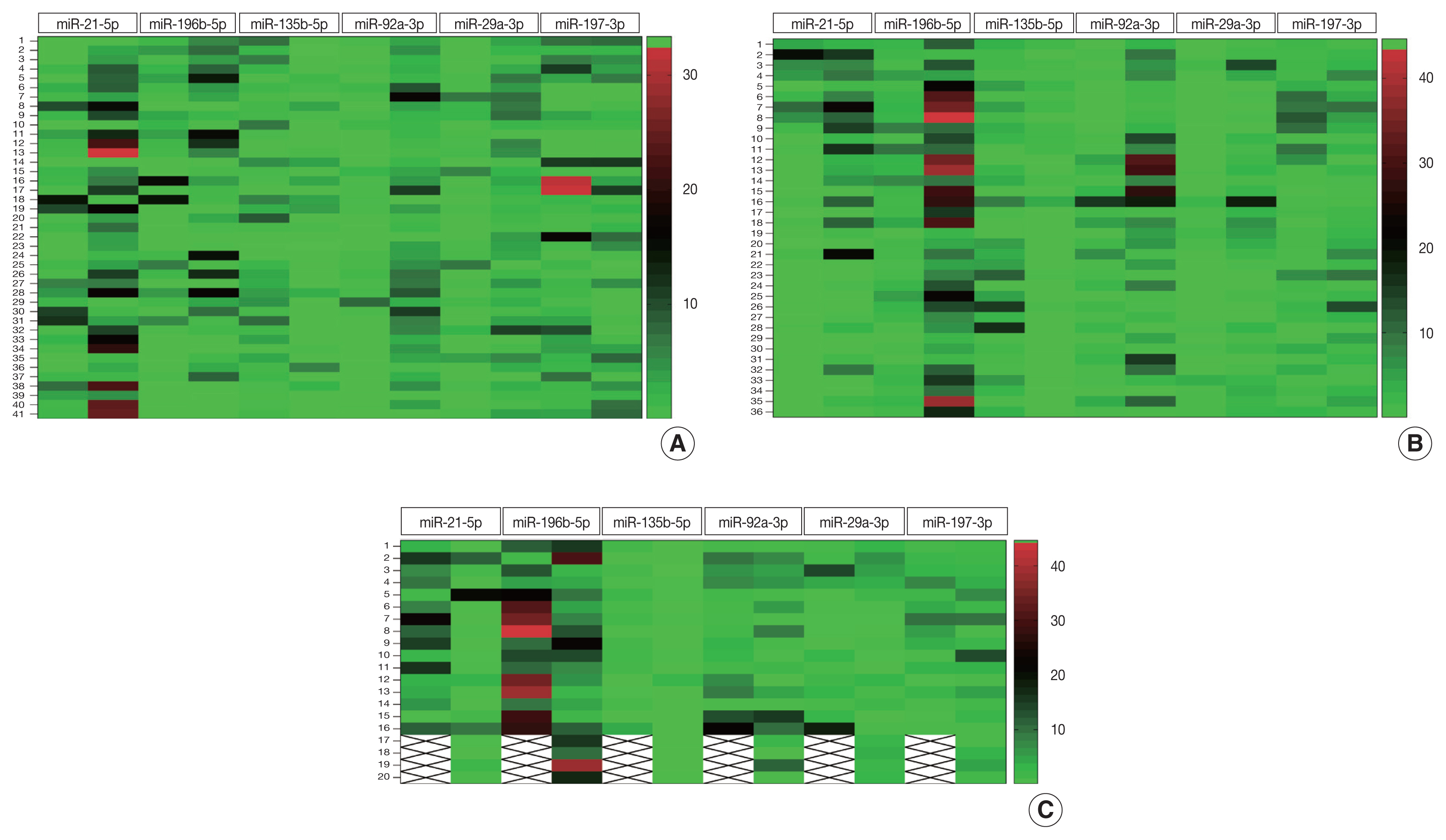
Using quantitative reverse transcription polymerase chain reaction (RT-qPCR), miR-21-5p, miR-29a-3p, miR-92a-3p, miR-135b-5p, miR-196b-5p, and miR-197-3p, expression was analyzed and compared between the malignant (n = 41) and the adjacent neoplasm free mucosal tissues (n = 41) of CRC patients. The findings were validated in plasma samples (n = 36) collected from the same CRC patients prior to surgery or any form of treatment and compared to plasma from their age and sex-matched controls (n = 36).
Results
MiR-21-5p, miR-29a-3p, miR-92a-3p, and miR- 196b-5p were upregulated and miR-135b-5p was downregulated in CRC malignant tissues compared to their expression in adjacent neoplasm-free tissue. This was further observed in the plasma of the same CRC cases compared to controls. MiR-92a-3p showed itself the most sensitive (0.93; p < .001) and most specific (0.95; p < .001) in detecting CRC in tissue. In plasma, miR-196b-5p was the most sensitive (0.97; p < .001) and specific (0.94; p < .001) in detecting CRC. Plasma miR-92a-3p and miR-196b-5p were the most sensitive (0.95; p < .001) and specific (0.94; p < .001) in the early detection of CRC.
Conclusions
Results show that specific miRNAs dysregulated in malignant tissues are released and can be detected in the circulation, supporting their potential as non-invasive biomarkers of CRC.
Keyword
Figure
Reference
-
References
1. Kavousipour S, Khademi F, Zamani M, Vakili B, Mokarram P. Novel biotechnology approaches in colorectal cancer diagnosis and therapy. Biotechnol Lett. 2017; 39:785–803.
Article2. Moreno CC, Mittal PK, Sullivan PS, et al. Colorectal cancer initial diagnosis: screening colonoscopy, diagnostic colonoscopy, or emergent surgery, and tumor stage and size at initial presentation. Clin Colorectal Cancer. 2016; 15:67–73.
Article3. Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021; 71:209–49.
Article4. Pawa N, Arulampalam T, Norton JD. Screening for colorectal cancer: established and emerging modalities. Nat Rev Gastroenterol Hepatol. 2011; 8:711–22.
Article5. Ahmed FE, Ahmed NC. MicroRNAs as molecular markers for colon cancer: diagnostic screening in stool and blood. Med Res Innov. 2017; 1:1–20.6. Ju J. miRNAs as biomarkers in colorectal cancer diagnosis and prognosis. Bioanalysis. 2010; 2:901–6.
Article7. Lin J, Chuang CC, Zuo L. Potential roles of microRNAs and ROS in colorectal cancer: diagnostic biomarkers and therapeutic targets. Oncotarget. 2017; 8:17328–46.
Article8. Nagy ZB, Wichmann B, Kalmar A, et al. Colorectal adenoma and carcinoma specific miRNA profiles in biopsy and their expression in plasma specimens. Clin Epigenetics. 2017; 9:22.
Article9. Ng EK, Chong WW, Jin H, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009; 58:1375–81.
Article10. Wikberg ML, Myte R, Palmqvist R, van Guelpen B, Ljuslinder I. Plasma miRNA can detect colorectal cancer, but how early? Cancer Med. 2018; 7:1697–705.11. Saini V, Dawar R, Suneja S, Gangopadhyay S, Kaur C. Can microRNA become next-generation tools in molecular diagnostics and therapeutics?: a systematic review. Egypt J Med Hum Genet. 2021; 22:4.
Article12. Buhagiar A, Seria E, Borg M, Borg J, Ayers D. Overview of microRNAs as liquid biopsy biomarkers for colorectal cancer sub-type profiling and chemoresistance. Cancer Drug Resist. 2021; 4:934–45.
Article13. Zuo Z, Jiang Y, Zeng S, et al. The value of microRNAs as the novel biomarkers for colorectal cancer diagnosis: a meta-analysis. Pathol Res Pract. 2020; 216:153130.
Article14. Yau TO, Tang CM, Harriss EK, Dickins B, Polytarchou C. Faecal microRNAs as a non-invasive tool in the diagnosis of colonic adenomas and colorectal cancer: a meta-analysis. Sci Rep. 2019; 9:9491.
Article15. Pardini B, Sabo AA, Birolo G, Calin GA. Noncoding RNAs in extracellular fluids as cancer biomarkers: the new frontier of liquid biopsies. Cancers (Basel). 2019; 11:1170.
Article16. Hibner G, Kimsa-Furdzik M, Francuz T. Relevance of microRNAs as potential diagnostic and prognostic markers in colorectal cancer. Int J Mol Sci. 2018; 19:2944.
Article17. Carter JV, Galbraith NJ, Yang D, Burton JF, Walker SP, Galandiuk S. Blood-based microRNAs as biomarkers for the diagnosis of colorectal cancer: a systematic review and meta-analysis. Br J Cancer. 2017; 116:762–74.
Article18. Yi R, Li Y, Wang FL, Miao G, Qi RM, Zhao YY. MicroRNAs as diagnostic and prognostic biomarkers in colorectal cancer. World J Gastrointest Oncol. 2016; 8:330–40.
Article19. Wang DD, Chen X, Yu DD, et al. miR-197: a novel biomarker for cancers. Gene. 2016; 591:313–9.
Article20. Zhi ML, Liu ZJ, Yi XY, Zhang LJ, Bao YX. Diagnostic performance of microRNA-29a for colorectal cancer: a meta-analysis. Genet Mol Res. 2015; 14:18018–25.
Article21. Zhang H, Li P, Ju H, et al. Diagnostic and prognostic value of microRNA-21 in colorectal cancer: an original study and individual participant data meta-analysis. Cancer Epidemiol Biomarkers Prev. 2014; 23:2783–92.
Article22. Yang X, Zeng Z, Hou Y, et al. MicroRNA-92a as a potential biomarker in diagnosis of colorectal cancer: a systematic review and meta-analysis. PLoS One. 2014; 9:e88745.
Article23. Motulsky HJ, Brown RE. Detecting outliers when fitting data with nonlinear regression: a new method based on robust nonlinear regression and the false discovery rate. BMC Bioinformatics. 2006; 7:123.24. Wu Y, Song Y, Xiong Y, et al. MicroRNA-21 (Mir-21) promotes cell growth and invasion by repressing tumor suppressor PTEN in colorectal cancer. Cell Physiol Biochem. 2017; 43:945–58.
Article25. Mima K, Nishihara R, Yang J, et al. MicroRNA MIR21 (miR-21) and PTGS2 expression in colorectal cancer and patient survival. Clin Cancer Res. 2016; 22:3841–8.26. Yu Y, Kanwar SS, Patel BB, et al. MicroRNA-21 induces stemness by downregulating transforming growth factor beta receptor 2 (TGFbetaR2) in colon cancer cells. Carcinogenesis. 2012; 33:68–76.
Article27. Feng YH, Tsao CJ. Emerging role of microRNA-21 in cancer. Biomed Rep. 2016; 5:395–402.
Article28. Yamada A, Horimatsu T, Okugawa Y, et al. Serum miR-21, miR-29a, and miR-125b are promising biomarkers for the early detection of colorectal neoplasia. Clin Cancer Res. 2015; 21:4234–42.
Article29. Bastaminejad S, Taherikalani M, Ghanbari R, Akbari A, Shabab N, Saidijam M. Investigation of microRNA-21 expression levels in serum and stool as a potential non-invasive biomarker for diagnosis of colorectal cancer. Iran Biomed J. 2017; 21:106–13.30. Almeida AL, Bernardes MV, Feitosa MR, et al. Serological under expression of microRNA-21, microRNA-34a and microRNA-126 in colorectal cancer. Acta Cir Bras. 2016; 31(Suppl 1):13–8.
Article31. Jurkovicova D, Smolkova B, Magyerkova M, et al. Down-regulation of traditional oncomiRs in plasma of breast cancer patients. Oncotarget. 2017; 8:77369–84.
Article32. Stiegelbauer V, Vychytilova-Faltejskova P, Karbiener M, et al. miR-196b-5p regulates colorectal cancer cell migration and metastases through interaction with HOXB7 and GALNT5. Clin Cancer Res. 2017; 23:5255–66.
Article33. Lu YC, Chang JT, Huang YC, et al. Combined determination of circulating miR-196a and miR-196b levels produces high sensitivity and specificity for early detection of oral cancer. Clin Biochem. 2015; 48:115–21.
Article34. Li X, Zhang G, Luo F, et al. Identification of aberrantly expressed miRNAs in rectal cancer. Oncol Rep. 2012; 28:77–84.
Article35. Wu J, Lin B, Yu S, et al. Exosomal miR-196b-5p is a potential diagnostic marker for colorectal cancer with metachronous liver metastasis. Transl Cancer Res. 2018; 7:1482–90.
Article36. Xu C, Gu L. The diagnostic effect of serum miR-196b as biomarker in colorectal cancer. Biomed Rep. 2017; 6:39–45.
Article37. Li Y, Zhang M, Chen H, et al. Ratio of miR-196s to HOXC8 messenger RNA correlates with breast cancer cell migration and metastasis. Cancer Res. 2010; 70:7894–904.
Article38. Braig S, Mueller DW, Rothhammer T, Bosserhoff AK. MicroRNA miR-196a is a central regulator of HOX-B7 and BMP4 expression in malignant melanoma. Cell Mol Life Sci. 2010; 67:3535–48.
Article39. Bhatia S, Kaul D, Varma N. Potential tumor suppressive function of miR-196b in B-cell lineage acute lymphoblastic leukemia. Mol Cell Biochem. 2010; 340:97–106.
Article40. Brunet Vega A, Pericay C, Moya I, et al. microRNA expression profile in stage III colorectal cancer: circulating miR-18a and miR-29a as promising biomarkers. Oncol Rep. 2013; 30:320–6.
Article41. Giraldez MD, Lozano JJ, Ramirez G, et al. Circulating microRNAs as biomarkers of colorectal cancer: results from a genome-wide profiling and validation study. Clin Gastroenterol Hepatol. 2013; 11:681–8.
Article42. Liu X, Lv X, Yang Q, Jin H, Zhou W, Fan Q. MicroRNA-29a functions as a tumor suppressor and increases cisplatin sensitivity by targeting NRAS in lung cancer. Technol Cancer Res Treat. 2018; 17:1533033818758905.
Article43. Tang W, Zhu Y, Gao J, et al. MicroRNA-29a promotes colorectal cancer metastasis by regulating matrix metalloproteinase 2 and E-cadherin via KLF4. Br J Cancer. 2014; 110:450–8.
Article44. Pei YF, Lei Y, Liu XQ. MiR-29a promotes cell proliferation and EMT in breast cancer by targeting ten eleven translocation 1. Biochim Biophys Acta. 2016; 1862:2177–85.
Article45. Trehoux S, Lahdaoui F, Delpu Y, et al. Micro-RNAs miR-29a and miR-330-5p function as tumor suppressors by targeting the MUC1 mucin in pancreatic cancer cells. Biochim Biophys Acta. 2015; 1853:2392–403.
Article46. Li M, Guan X, Sun Y, et al. miR-92a family and their target genes in tumorigenesis and metastasis. Exp Cell Res. 2014; 323:1–6.
Article47. Tsuchida A, Ohno S, Wu W, et al. miR-92 is a key oncogenic component of the miR-17-92 cluster in colon cancer. Cancer Sci. 2011; 102:2264–71.
Article48. Ke TW, Wei PL, Yeh KT, Chen WT, Cheng YW. MiR-92a promotes cell metastasis of colorectal cancer through PTEN-mediated PI3K/AKT pathway. Ann Surg Oncol. 2015; 22:2649–55.
Article49. Li J, Liang H, Bai M, et al. miR-135b promotes cancer progression by targeting transforming growth factor beta receptor II (TGFBR2) in colorectal cancer. PLoS One. 2015; 10:e0130194.
Article50. Magalhaes L, Quintana LG, Lopes DCF, et al. APC gene is modulated by hsa-miR-135b-5p in both diffuse and intestinal gastric cancer subtypes. BMC Cancer. 2018; 18:1055.
Article51. Zekri AR, Youssef AS, Lotfy MM, et al. Circulating serum miRNAs as diagnostic markers for colorectal cancer. PLoS One. 2016; 11:e0154130.
Article52. Bastaminejad S, Taherikalani M, Ghanbari R, et al. Serum and stool miR-135b levels as a potential diagnostic biomarker for colorectal cancer. Clin Exp Invest. 2020; 1:1–6.53. Uddin MN, Li M, Wang X. Identification of transcriptional markers and microRNA-mRNA regulatory networks in colon cancer by integrative analysis of mRNA and microRNA expression profiles in colon tumor stroma. Cells. 2019; 8:1054.
Article54. Reichholf B, Herzog VA, Fasching N, Manzenreither RA, Sowemimo I, Ameres SL. Time-resolved small RNA sequencing unravels the molecular principles of microRNA homeostasis. Mol Cell. 2019; 75:756–68.
Article55. Cojocneanu R, Braicu C, Raduly L, et al. Plasma and tissue specific miRNA expression pattern and functional analysis associated to colorectal cancer patients. Cancers (Basel). 2020; 12:843.
Article56. Nagy ZB, Bartak BK, Kalmar A, et al. Comparison of circulating miRNAs expression alterations in matched tissue and plasma samples during colorectal cancer progression. Pathol Oncol Res. 2019; 25:97–105.
Article57. Ma R, Jiang T, Kang X. Circulating microRNAs in cancer: origin, function and application. J Exp Clin Cancer Res. 2012; 31:38.
Article58. De Rosa M, Rega D, Costabile V, et al. The biological complexity of colorectal cancer: insights into biomarkers for early detection and personalized care. Therap Adv Gastroenterol. 2016; 9:861–86.
Article59. Balboa E, Carracedo A, Barros F. The complexity of colorectal cancer biology: putting bricks on the path to personalized medicine. Colorectal cancer. Khan JS, editor. Rijeka: Intech;2014. p. 434–66.60. Nguyen HT, Duong HQ. The molecular characteristics of colorectal cancer: implications for diagnosis and therapy. Oncol Lett. 2018; 16:9–18.61. Diaz-Cano SJ. Tumor heterogeneity: mechanisms and bases for a reliable application of molecular marker design. Int J Mol Sci. 2012; 13:1951–2011.
Article62. Buikhuisen JY, Torang A, Medema JP. Exploring and modelling colon cancer inter-tumour heterogeneity: opportunities and challenges. Oncogenesis. 2020; 9:66.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Role of MicroRNAs in Colorectal Cancer
- Expression of Inducible Nitric Oxide Synthase in Human Colorectal
- Cell-Free miR-27a, a Potential Diagnostic and Prognostic Biomarker for Gastric Cancer
- Clinical Aspect of MicroRNA in Lung Cancer
- Effects of β-carotene on Expression of Selected MicroRNAs, Histone Acetylation, and DNA Methylation in Colon Cancer Stem Cells