Cancer Res Treat.
2023 Apr;55(2):452-467. 10.4143/crt.2022.910.
Cancer-Specific Sequences in the Diagnosis and Treatment of NUT Carcinoma
- Affiliations
-
- 1Department of Health Sciences and Technology, SAIHST, Sungkyunkwan University, Seoul, Korea
- 2Laboratory of Molecular Pathology and Theranostics, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
- 3Department of Digital Health, SAIHST, Sungkyunkwan University, Seoul, Korea
- 4Department of Otolaryngology, Ajou University School of Medicine, Suwon, Korea
- 5Department of Pathology, Seoul National University College of Medicine, Seoul, Korea
- 6Cancer Research institute, Seoul National University, Seoul, Korea
- 7Laboratory of Molecular Pathology and Cancer Genomics, Research Institute of Pharmaceutical Sciences and College of Pharmacy, Seoul National University, Seoul, Korea
- 8Bio-Max/N-Bio, Seoul National University, Seoul, Korea
- 9Division of Hematology-Oncology, Department of Medicine, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
- 10Department of Pathology and Translational Genomics, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
- KMID: 2541232
- DOI: http://doi.org/10.4143/crt.2022.910
Abstract
- Purpose
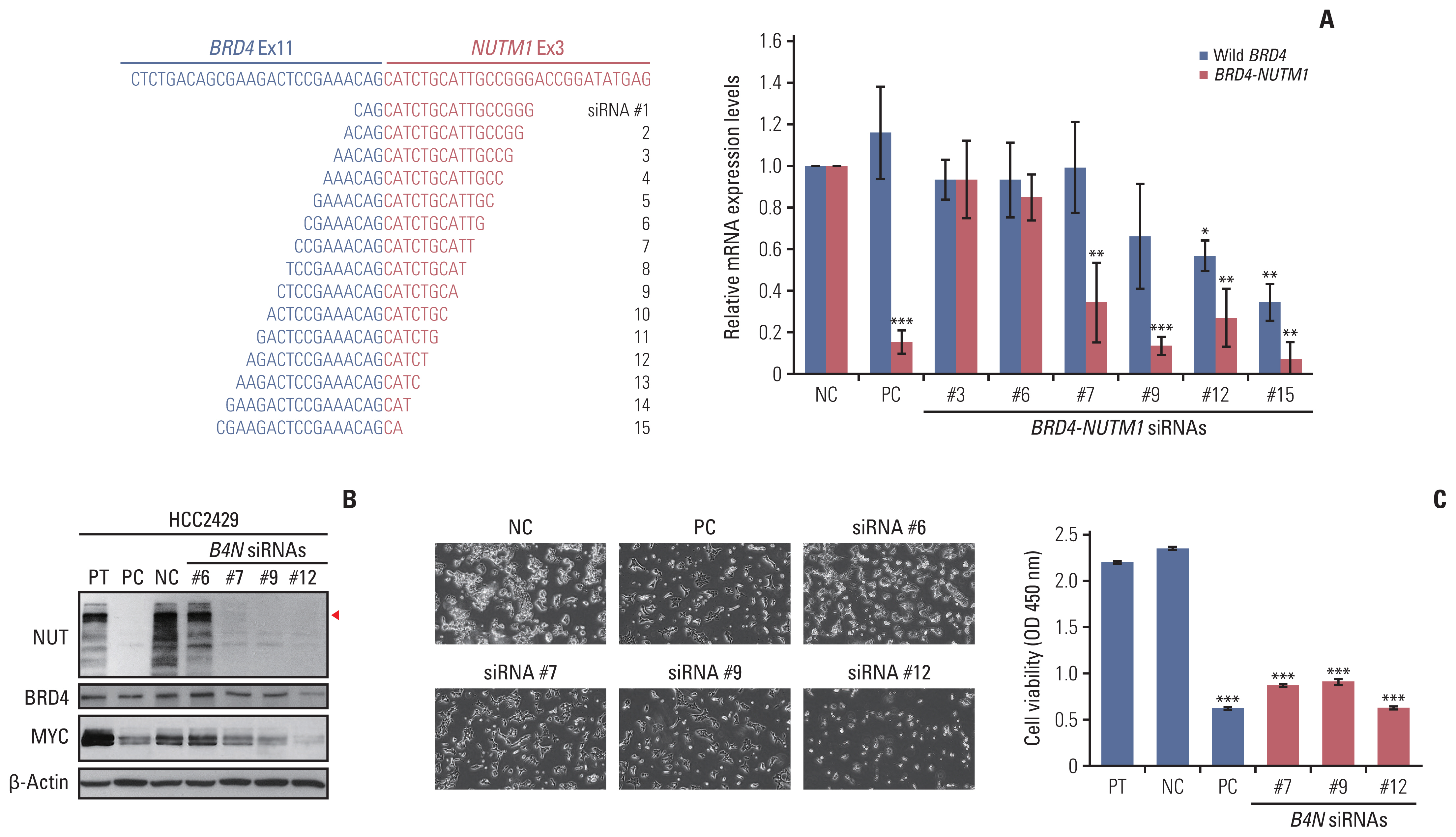
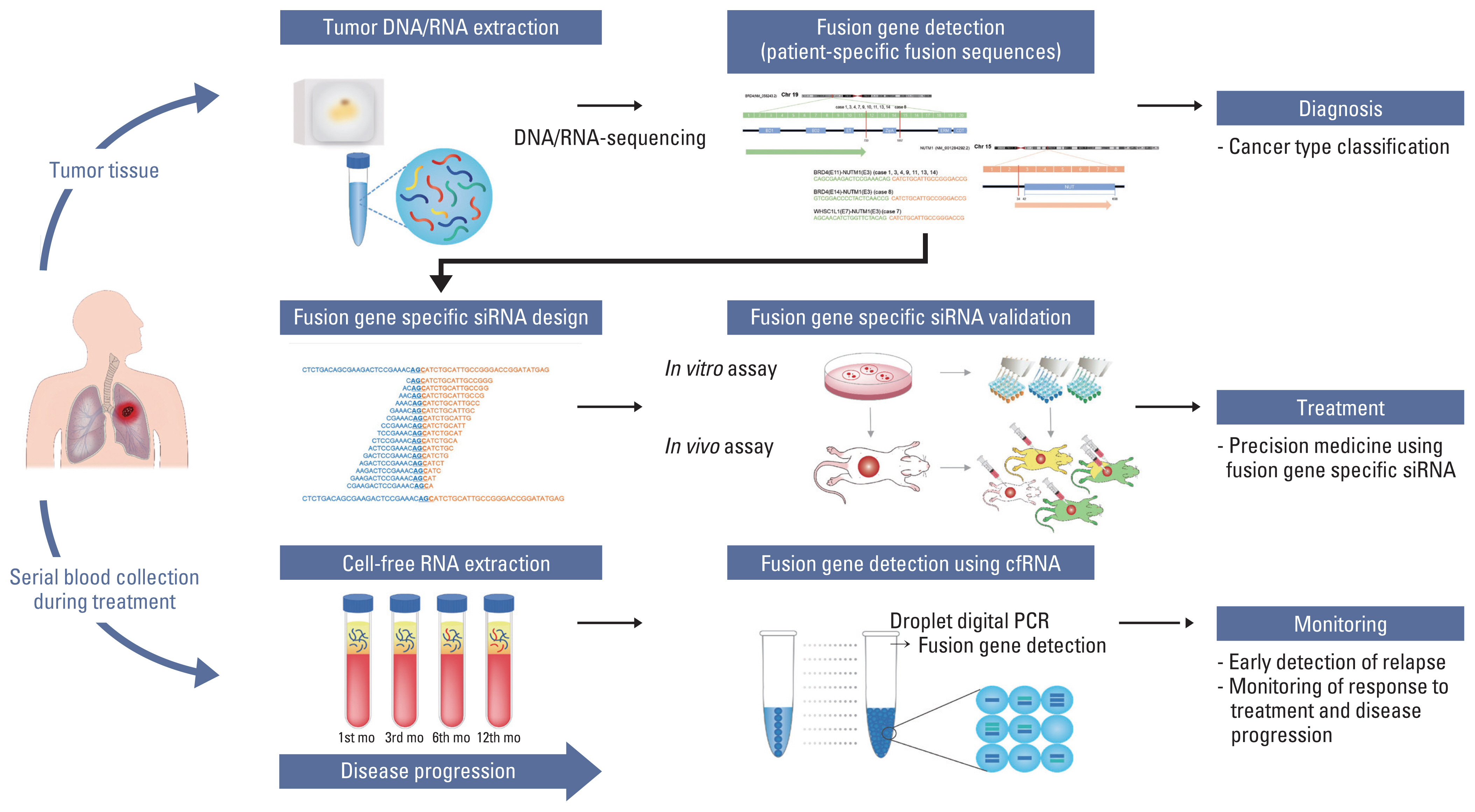
NUT carcinoma (NC) is a solid tumor caused by the rearrangement of NUTM1 that usually develops in midline structures, such as the thorax. No standard treatment has been established despite high lethality. Thus, we investigated whether targeting the junction region of NUTM1 fusion breakpoints could serve as a potential treatment option for NC.
Materials and Methods
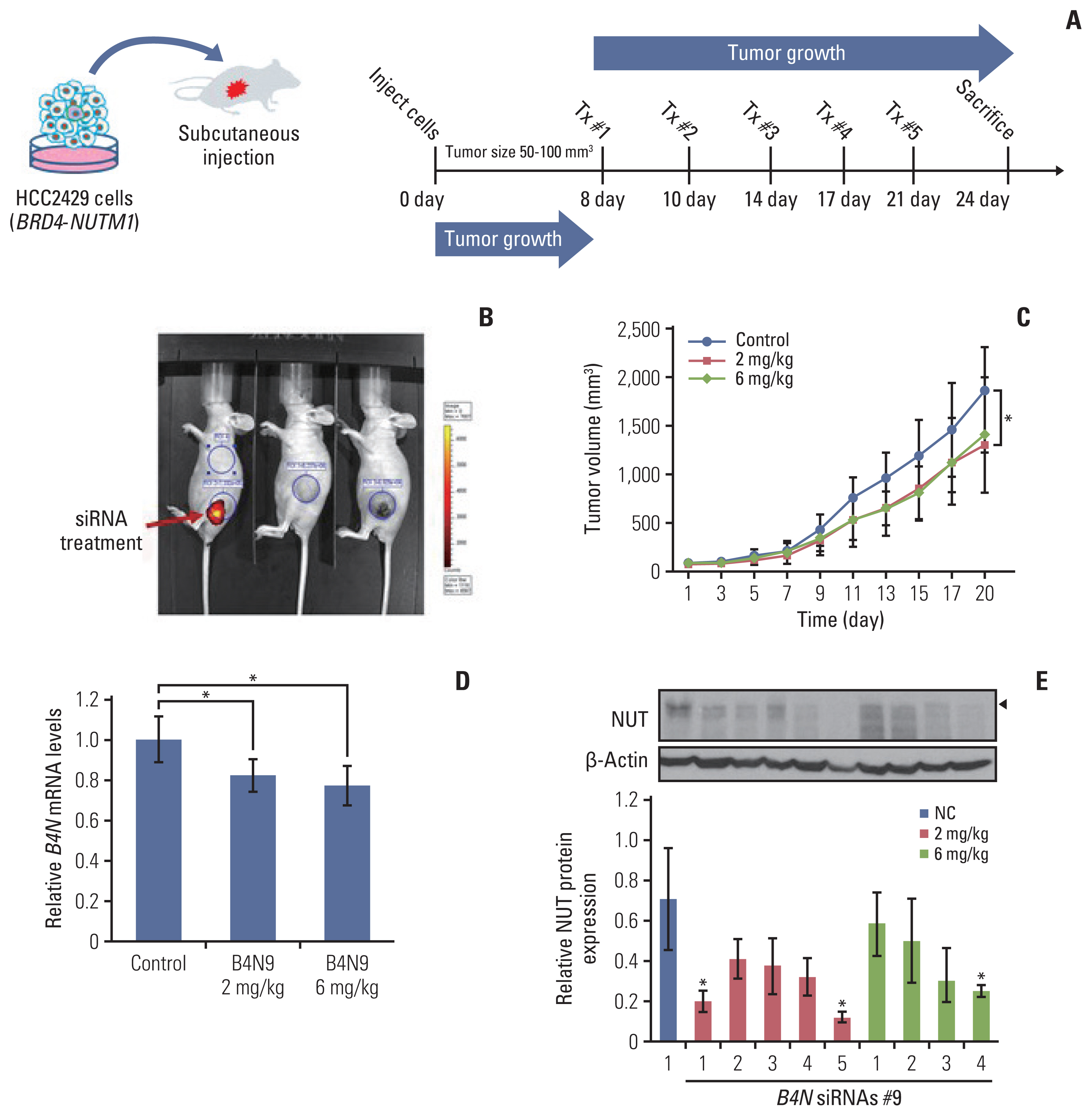
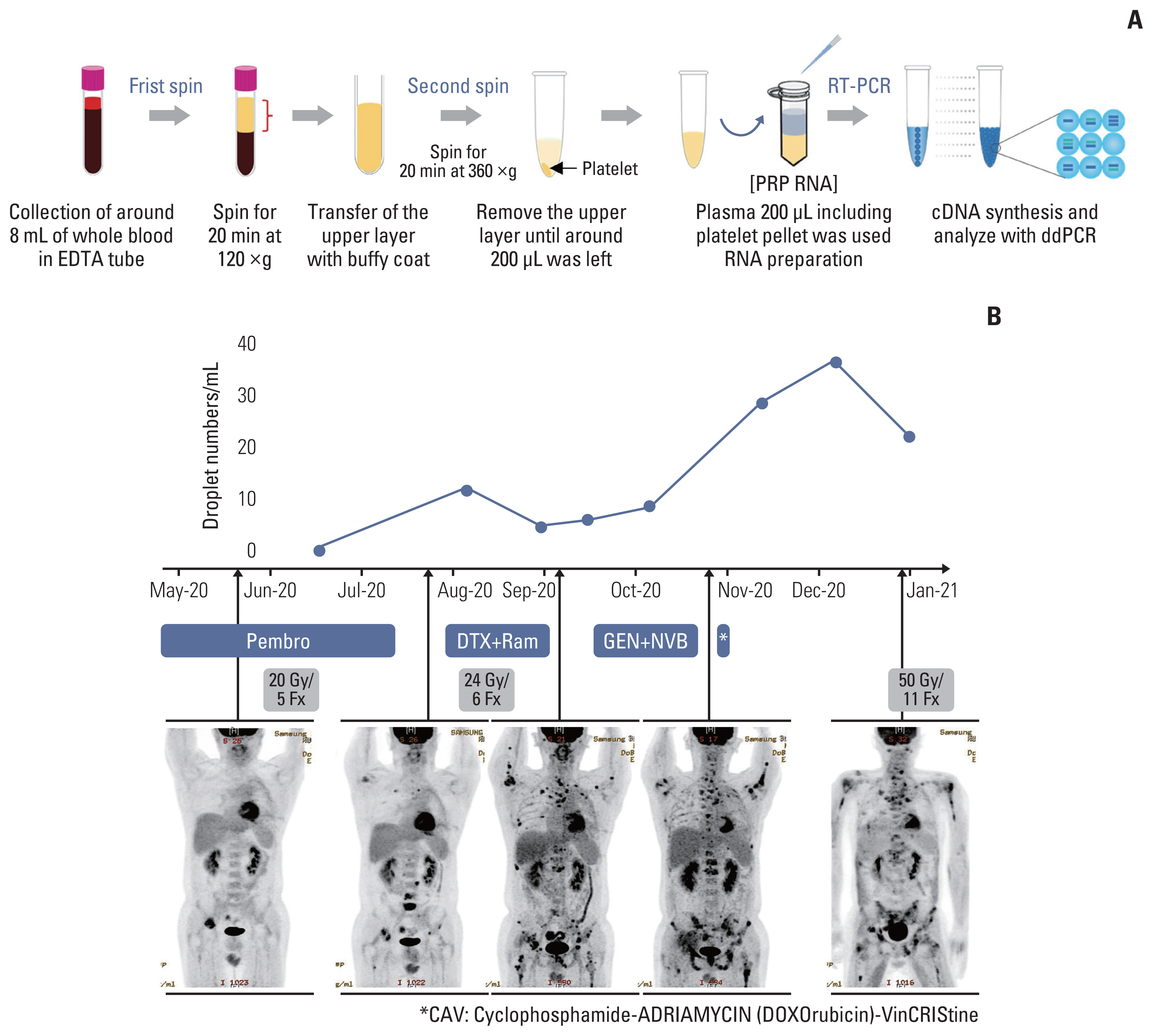
We designed and evaluated a series of small interfering RNAs (siRNAs) targeting the junction region of BRD4-NUTM1 fusion (B4N), the most common form of NUTM1 fusion. Droplet digital polymerase chain reaction using the blood of patients was also tested to evaluate the treatment responses by the junction sequence of the B4N fusion transcripts.
Results
As expected, the majority of NC fusion types were B4N (12 of 18, 67%). B4N fusion-specific siRNA treatment on NC cells showed specific inhibitory effects on the B4N fusion transcript and fusion protein without affecting the endogenous expression of the parent genes, resulting in decreased relative cell growth and attenuation of tumor size. In addition, the fusion transcript levels in platelet-rich-plasma samples of the NC patients with systemic metastasis showed a negative correlation with therapeutic effect, suggesting its potential as a measure of treatment responsiveness.
Conclusion
This study suggests that tumor-specific sequences could be used to treat patients with fusion genes as part of precision medicine for a rare but deadly disease.
Figure
Reference
-
References
1. French CA. NUT carcinoma: clinicopathologic features, pathogenesis, and treatment. Pathol Int. 2018; 68:583–95.
Article2. Jung M, Kim S, Lee JK, Yoon SO, Park HS, Hong SW, et al. Clinicopathological and preclinical findings of NUT carcinoma: a multicenter study. Oncologist. 2019; 24:e740–8.
Article3. Parikh SA, French CA, Costello BA, Marks RS, Dronca RS, Nerby CL, et al. NUT midline carcinoma: an aggressive intra-thoracic neoplasm. J Thorac Oncol. 2013; 8:1335–8.
Article4. Chau NG, Ma C, Danga K, Al-Sayegh H, Nardi V, Barrette R, et al. An anatomical site and genetic-based prognostic model for patients with nuclear protein in testis (NUT) midline carcinoma: analysis of 124 patients. JNCI Cancer Spectr. 2020. 4:pkz094.
Article5. Chau NG, Hurwitz S, Mitchell CM, Aserlind A, Grunfeld N, Kaplan L, et al. Intensive treatment and survival outcomes in NUT midline carcinoma of the head and neck. Cancer. 2016; 122:3632–40.6. Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, et al. The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015; 10:1243–60.7. Mao N, Liao Z, Wu J, Liang K, Wang S, Qin S, et al. Diagnosis of NUT carcinoma of lung origin by next-generation sequencing: case report and review of the literature. Cancer Biol Ther. 2019; 20:150–6.8. Donati B, Lorenzini E, Ciarrocchi A. BRD4 and cancer: going beyond transcriptional regulation. Mol Cancer. 2018; 17:164.9. Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011; 146:904–17.10. Grayson AR, Walsh EM, Cameron MJ, Godec J, Ashworth T, Ambrose JM, et al. MYC, a downstream target of BRD-NUT, is necessary and sufficient for the blockade of differentiation in NUT midline carcinoma. Oncogene. 2014; 33:1736–42.11. French CA, Miyoshi I, Kubonishi I, Grier HE, Perez-Atayde AR, Fletcher JA. BRD4-NUT fusion oncogene: a novel mechanism in aggressive carcinoma. Cancer Res. 2003; 63:304–7.12. Shiota H, Barral S, Buchou T, Tan M, Coute Y, Charbonnier G, et al. Nut directs p300-dependent, genome-wide H4 hyperacetylation in male germ cells. Cell Rep. 2018; 24:3477–87.13. Eagen KP, French CA. Supercharging BRD4 with NUT in carcinoma. Oncogene. 2021; 40:1396–408.14. Schwartz BE, Hofer MD, Lemieux ME, Bauer DE, Cameron MJ, West NH, et al. Differentiation of NUT midline carcinoma by epigenomic reprogramming. Cancer Res. 2011; 71:2686–96.15. Alekseyenko AA, Walsh EM, Zee BM, Pakozdi T, Hsi P, Lemieux ME, et al. Ectopic protein interactions within BRD4-chromatin complexes drive oncogenic megadomain formation in NUT midline carcinoma. Proc Natl Acad Sci USA. 2017; 114:E4184–92.16. Ameratunga M, Brana I, Bono P, Postel-Vinay S, Plummer R, Aspegren J, et al. First-in-human phase 1 open label study of the BET inhibitor ODM-207 in patients with selected solid tumours. Br J Cancer. 2020; 123:1730–6.17. French CA. Small-molecule targeting of BET proteins in cancer. Adv Cancer Res. 2016; 131:21–58.18. Stathis A, Zucca E, Bekradda M, Gomez-Roca C, Delord JP, de La Motte Rouge T, et al. Clinical response of carcinomas harboring the BRD4-NUT oncoprotein to the targeted bromodomain inhibitor OTX015/MK-8628. Cancer Discov. 2016; 6:492–500.19. Lewin J, Soria JC, Stathis A, Delord JP, Peters S, Awada A, et al. Phase Ib trial with birabresib, a small-molecule inhibitor of bromodomain and extraterminal proteins, in patients with selected advanced solid tumors. J Clin Oncol. 2018; 36:3007–14.20. Piha-Paul SA, Hann CL, French CA, Cousin S, Brana I, Cassier PA, et al. Phase 1 study of molibresib (GSK525762), a bromodomain and extra-terminal domain protein inhibitor, in NUT carcinoma and other solid tumors. JNCI Cancer Spectr. 2020; 4:pkz093.21. Subhan MA, Torchilin VP. Efficient nanocarriers of siRNA therapeutics for cancer treatment. Transl Res. 2019; 214:62–91.22. Hu B, Zhong L, Weng Y, Peng L, Huang Y, Zhao Y, et al. Therapeutic siRNA: state of the art. Signal Transduct Target Ther. 2020; 5:101.23. Gavrilov K, Seo YE, Tietjen GT, Cui J, Cheng CJ, Saltzman WM. Enhancing potency of siRNA targeting fusion genes by optimization outside of target sequence. Proc Natl Acad Sci USA. 2015; 112:E6597–605.24. Parker Kerrigan BC, Ledbetter D, Kronowitz M, Phillips L, Gumin J, Hossain A, et al. RNAi technology targeting the FGFR3-TACC3 fusion breakpoint: an opportunity for precision medicine. Neurooncol Adv. 2020. 2:vdaa132.25. Lee JK, Louzada S, An Y, Kim SY, Kim S, Youk J, et al. Complex chromosomal rearrangements by single catastrophic pathogenesis in NUT midline carcinoma. Ann Oncol. 2017; 28:890–7.26. Cho YA, Choi YL, Hwang I, Lee K, Cho JH, Han J. Clinicopathological characteristics of primary lung nuclear protein in testis carcinoma: a single-institute experience of 10 cases. Thorac Cancer. 2020; 11:3205–12.27. Amary MF, Berisha F, Bernardi Fdel C, Herbert A, James M, Reis-Filho JS, et al. Detection of SS18-SSX fusion transcripts in formalin-fixed paraffin-embedded neoplasms: analysis of conventional RT-PCR, qRT-PCR and dual color FISH as diagnostic tools for synovial sarcoma. Mod Pathol. 2007; 20:482–96.28. McEvoy CR, Holliday H, Thio N, Mitchell C, Choong DY, Yellapu B, et al. A MXI1-NUTM1 fusion protein with MYC-like activity suggests a novel oncogenic mechanism in a subset of NUTM1-rearranged tumors. Lab Invest. 2021; 101:26–37.29. Le Loarer F, Pissaloux D, Watson S, Godfraind C, Galmiche-Rolland L, Silva K, et al. Clinicopathologic features of CIC-NUTM1 sarcomas, a new molecular variant of the family of CIC-fused sarcomas. Am J Surg Pathol. 2019; 43:268–76.30. Dickson BC, Sung YS, Rosenblum MK, Reuter VE, Harb M, Wunder JS, et al. NUTM1 gene fusions characterize a subset of undifferentiated soft tissue and visceral tumors. Am J Surg Pathol. 2018; 42:636–45.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A Case Report of 4-Years Old Patient With Nuclear Protein in Testis Midline Carcinoma of Larynx
- Two Cases of Nuclear Protein in Testis Midline Carcinomas of Sinonasal Tract
- Cashew nut allergy in Singaporean children
- Radiologic Manifestations of Pulmonary Nuclear Protein in Testis Midline Carcinoma: A Case Report
- Abrupt Dyskeratotic and Squamoid Cells in Poorly Differentiated Carcinoma: Case Study of Two Thoracic NUT Midline Carcinomas with Cytohistologic Correlation