Diabetes Metab J.
2023 Mar;47(2):255-266. 10.4093/dmj.2021.0375.
Genome-Wide Association Study on Longitudinal Change in Fasting Plasma Glucose in Korean Population
- Affiliations
-
- 1Institute of Health and Environment, Seoul National University, Seoul, Korea
- 2Department of Internal Medicine, Seoul National University Hospital, Seoul, Korea
- 3Department of Internal Medicine, Seoul National University Hospital Healthcare System Gangnam Center, Seoul, Korea
- 4Department of Bioconvergence & Engineering, Dankook University, Yongin, Korea
- 5Department of Internal Medicine, Seoul National University College of Medicine, Seoul, Korea
- 6Department of Molecular Medicine and Biopharmaceutical Sciences, Graduate School of Convergence Science and Technology, Seoul National University, Seoul, Korea
- 7Department of Public Health Sciences, Seoul National University, Seoul, Korea
- 8RexSoft Inc., Seoul, Korea
- 9Department of Preventive Medicine, Ajou University School of Medicine, Suwon, Korea
- KMID: 2540522
- DOI: http://doi.org/10.4093/dmj.2021.0375
Abstract
- Background
Genome-wide association studies (GWAS) on type 2 diabetes mellitus (T2DM) have identified more than 400 distinct genetic loci associated with diabetes and nearly 120 loci for fasting plasma glucose (FPG) and fasting insulin level to date. However, genetic risk factors for the longitudinal deterioration of FPG have not been thoroughly evaluated. We aimed to identify genetic variants associated with longitudinal change of FPG over time.
Methods
We used two prospective cohorts in Korean population, which included a total of 10,528 individuals without T2DM. GWAS of repeated measure of FPG using linear mixed model was performed to investigate the interaction of genetic variants and time, and meta-analysis was conducted. Genome-wide complex trait analysis was used for heritability calculation. In addition, expression quantitative trait loci (eQTL) analysis was performed using the Genotype-Tissue Expression project.
Results
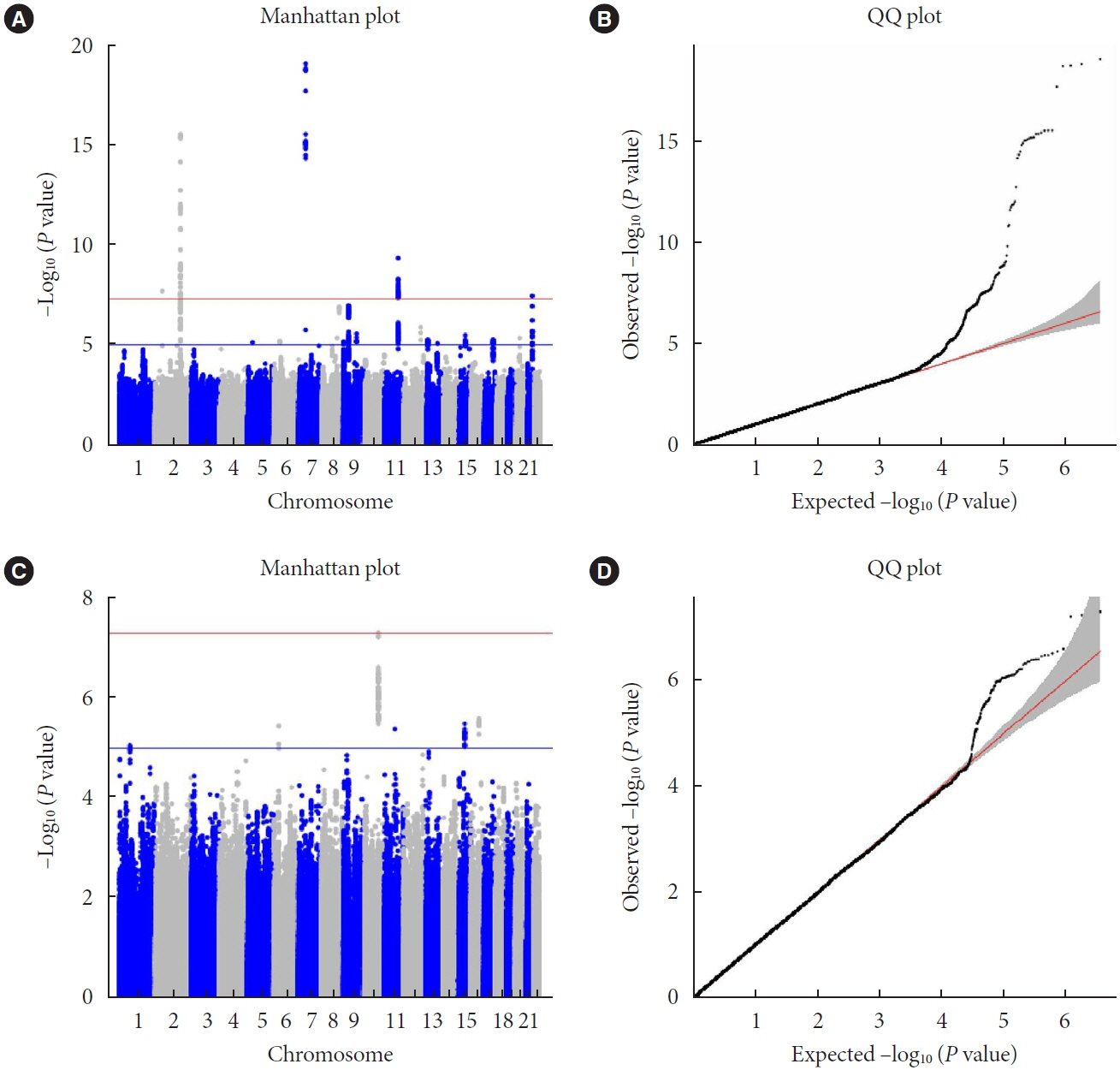
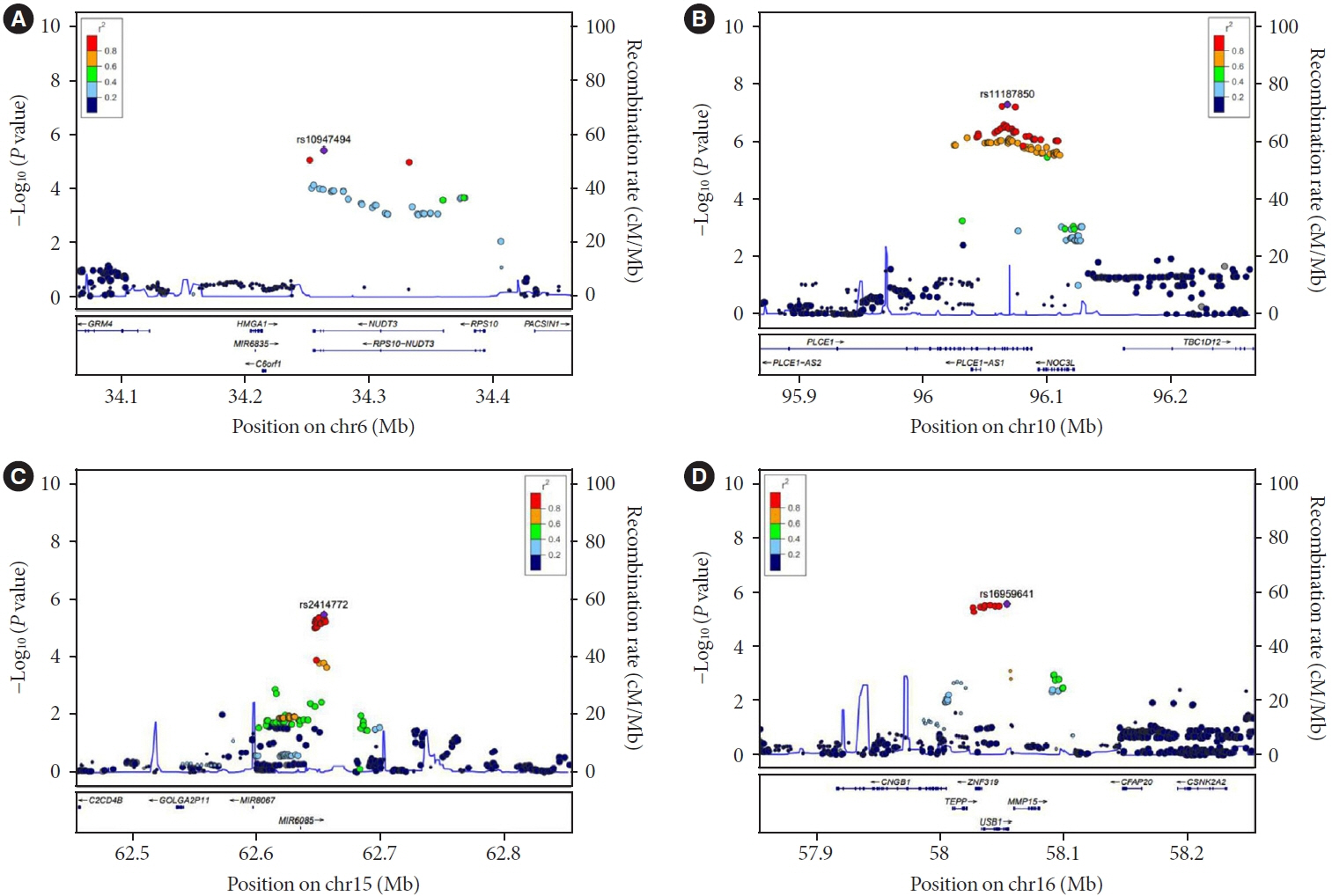
A small portion (4%) of the genome-wide single nucleotide polymorphism (SNP) interaction with time explained the total phenotypic variance of longitudinal change in FPG. A total of four known genetic variants of FPG were associated with repeated measure of FPG levels. One SNP (rs11187850) showed a genome-wide significant association for genetic interaction with time. The variant is an eQTL for NOC3 like DNA replication regulator (NOC3L) gene in pancreas and adipose tissue. Furthermore, NOC3L is also differentially expressed in pancreatic β-cells between subjects with or without T2DM. However, this variant was not associated with increased risk of T2DM nor elevated FPG level.
Conclusion
We identified rs11187850, which is an eQTL of NOC3L, to be associated with longitudinal change of FPG in Korean population.
Figure
Reference
-
1. Tabak AG, Jokela M, Akbaraly TN, Brunner EJ, Kivimaki M, Witte DR. Trajectories of glycaemia, insulin sensitivity, and insulin secretion before diagnosis of type 2 diabetes: an analysis from the Whitehall II study. Lancet. 2009; 373:2215–21.2. Heianza Y, Arase Y, Fujihara K, Hsieh SD, Saito K, Tsuji H, et al. Longitudinal trajectories of HbA1c and fasting plasma glucose levels during the development of type 2 diabetes: the Toranomon Hospital Health Management Center Study 7 (TOPICS 7). Diabetes Care. 2012; 35:1050–2.3. Meigs JB, Muller DC, Nathan DM, Blake DR, Andres R; Baltimore Longitudinal Study of Aging. The natural history of progression from normal glucose tolerance to type 2 diabetes in the Baltimore Longitudinal Study of Aging. Diabetes. 2003; 52:1475–84.4. Chia CW, Egan JM, Ferrucci L. Age-related changes in glucose metabolism, hyperglycemia, and cardiovascular risk. Circ Res. 2018; 123:886–904.5. Zhang X, Gregg EW, Williamson DF, Barker LE, Thomas W, Bullard KM, et al. A1C level and future risk of diabetes: a systematic review. Diabetes Care. 2010; 33:1665–73.6. Mahajan A, Taliun D, Thurner M, Robertson NR, Torres JM, Rayner NW, et al. Fine-mapping type 2 diabetes loci to singlevariant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018; 50:1505–13.7. Flannick J, Mercader JM, Fuchsberger C, Udler MS, Mahajan A, Wessel J, et al. Exome sequencing of 20,791 cases of type 2 diabetes and 24,440 controls. Nature. 2019; 570:71–6.8. Liu CT, Raghavan S, Maruthur N, Kabagambe EK, Hong J, Ng MC, et al. Trans-ethnic meta-analysis and functional annotation illuminates the genetic architecture of fasting glucose and insulin. Am J Hum Genet. 2016; 99:56–75.9. Sikorska K, Rivadeneira F, Groenen PJ, Hofman A, Uitterlinden AG, Eilers PH, et al. Fast linear mixed model computations for genome-wide association studies with longitudinal data. Stat Med. 2013; 32:165–80.10. Little RJA, Rubin DB. Statistical analysis with missing data. 3rd ed. Hoboken: John Wiley & Sons;2020.11. Ohn JH, Kwak SH, Cho YM, Lim S, Jang HC, Park KS, et al. 10-Year trajectory of β-cell function and insulin sensitivity in the development of type 2 diabetes: a community-based prospective cohort study. Lancet Diabetes Endocrinol. 2016; 4:27–34.12. Moon S, Kim YJ, Han S, Hwang MY, Shin DM, Park MY, et al. The Korea Biobank Array: design and identification of coding variants associated with blood biochemical traits. Sci Rep. 2019; 9:1382.13. Seo S, Park K, Lee JJ, Choi KY, Lee KH, Won S. SNP genotype calling and quality control for multi-batch-based studies. Genes Genomics. 2019; 41:927–39.14. Song YE, Lee S, Park K, Elston RC, Yang HJ, Won S. ONETOOL for the analysis of family-based big data. Bioinformatics. 2018; 34:2851–3.15. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010; 38:e164.16. Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009; 5:e1000529.17. Team RC. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing;2013.18. Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient metaanalysis of genomewide association scans. Bioinformatics. 2010; 26:2190–1.19. Bacanu SA, Devlin B, Roeder K. The power of genomic control. Am J Hum Genet. 2000; 66:1933–44.20. Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011; 88:76–82.21. GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013; 45:580–5.22. Dominguez V, Raimondi C, Somanath S, Bugliani M, Loder MK, Edling CE, et al. Class II phosphoinositide 3-kinase regulates exocytosis of insulin granules in pancreatic beta cells. J Biol Chem. 2011; 286:4216–25.23. Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014; 46:310–5.24. Bland JM, Altman DG. Multiple significance tests: the Bonferroni method. BMJ. 1995; 310:170.25. Gauderman WJ. Sample size requirements for matched casecontrol studies of gene-environment interaction. Stat Med. 2002; 21:35–50.26. Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010; 42:105–16.27. Kim YJ, Go MJ, Hu C, Hong CB, Kim YK, Lee JY, et al. Largescale genome-wide association studies in East Asians identify new genetic loci influencing metabolic traits. Nat Genet. 2011; 43:990–5.28. Li X, Shu YH, Xiang AH, Trigo E, Kuusisto J, Hartiala J, et al. Additive effects of genetic variation in GCK and G6PC2 on insulin secretion and fasting glucose. Diabetes. 2009; 58:2946–53.29. Prokopenko I, Langenberg C, Florez JC, Saxena R, Soranzo N, Thorleifsson G, et al. Variants in MTNR1B influence fasting glucose levels. Nat Genet. 2009; 41:77–81.30. Hu C, Zhang R, Wang C, Yu W, Lu J, Ma X, et al. Effects of GCK, GCKR, G6PC2 and MTNR1B variants on glucose metabolism and insulin secretion. PLoS One. 2010; 5:e11761.31. Furlotte NA, Eskin E, Eyheramendy S. Genome-wide association mapping with longitudinal data. Genet Epidemiol. 2012; 36:463–71.32. Evangelou E, Warren HR, Mosen-Ansorena D, Mifsud B, Pazoki R, Gao H, et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet. 2018; 50:1412–25.33. Gazal S, Loh PR, Finucane HK, Ganna A, Schoech A, Sunyaev S, et al. Functional architecture of low-frequency variants highlights strength of negative selection across coding and non-coding annotations. Nat Genet. 2018; 50:1600–7.34. Zhu Z, Wang X, Li X, Lin Y, Shen S, Liu CL, et al. Genetic overlap of chronic obstructive pulmonary disease and cardiovascular disease-related traits: a large-scale genome-wide cross-trait analysis. Respir Res. 2019; 20:64.35. Pirruccello JP, Chaffin MD, Chou EL, Fleming SJ, Lin H, Nekoui M, et al. Deep learning enables genetic analysis of the human thoracic aorta. Nat Genet. 2022; 54:40–51.36. Johmura Y, Watanabe K, Kishimoto K, Ueda T, Shimada S, Osada S, et al. Fad24 causes hyperplasia in adipose tissue and improves glucose metabolism. Biol Pharm Bull. 2009; 32:1656–64.37. Tominaga K, Johmura Y, Nishizuka M, Imagawa M. Fad24, a mammalian homolog of Noc3p, is a positive regulator in adipocyte differentiation. J Cell Sci. 2004; 117(Pt 25)(Pt 25):6217–26.38. Liu CT, Merino J, Rybin D, DiCorpo D, Benke KS, BraggGresham JL, et al. Genome-wide association study of change in fasting glucose over time in 13,807 non-diabetic European ancestry individuals. Sci Rep. 2019; 9:9439.39. Kwak SH, Chae J, Lee S, Choi S, Koo BK, Yoon JW, et al. Nonsynonymous variants in PAX4 and GLP1R are associated with type 2 diabetes in an East Asian population. Diabetes. 2018; 67:1892–902.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Replication of Interactions between Genome-Wide Genetic Variants and Body Mass Index in Fasting Glucose and Insulin Levels
- A Short History of the Genome-Wide Association Study: Where We Were and Where We Are Going
- Genome-Wide Association Study Identifies Two Novel Loci with Sex-Specific Effects for Type 2 Diabetes Mellitus and Glycemic Traits in a Korean Population
- The Appropriteness of New ADA Diagnostin Criteria for Diabetes Mellitus in Korean Population
- Genome-Wide Association Study of Hepatitis in Korean Populations