Int J Stem Cells.
2023 Feb;16(1):36-43. 10.15283/ijsc22177.
Generation of Induced Pluripotent Stem Cells from Lymphoblastoid Cell Lines by Electroporation of Episomal Vectors
- Affiliations
-
- 1Department of Medical Life Sciences, College of Medicine, The Catholic University of Korea, Seoul, Korea
- 2School of Biotechnology and Healthcare, Wuyi University, Jiangmen, China
- 3Department of General Pediatrics, University of Children’s Hospital Muenster, Muenster, Germany
- 4Department of Cell and Developmental Biology, Max Planck Institute for Molecular Biomedicine, Münster, Germany
- 5Department of Cardiac Development and Remodelling, Max-Planck-Institute for Heart and Lung Research, Bad Nauheim, Germany
- KMID: 2539619
- DOI: http://doi.org/10.15283/ijsc22177
Abstract
- Background and Objectives
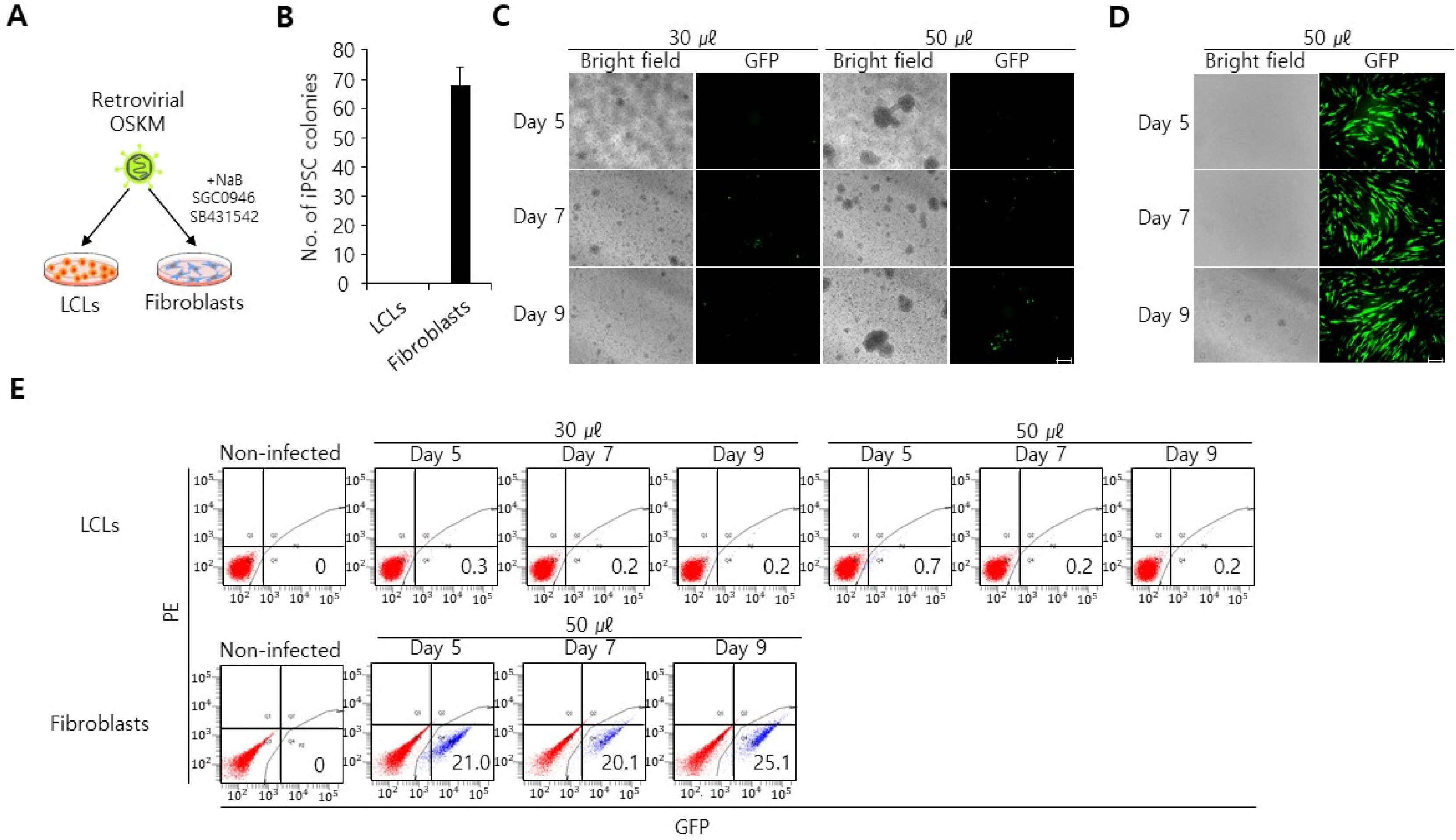
Lymphoblastoid cell lines (LCLs) deposited from disease-affected individuals could be a valuable donor cell source for generating disease-specific induced pluripotent stem cells (iPSCs). However, generation of iPSCs from the LCLs is still challenging, as yet no effective gene delivery strategy has been developed.
Methods and Results
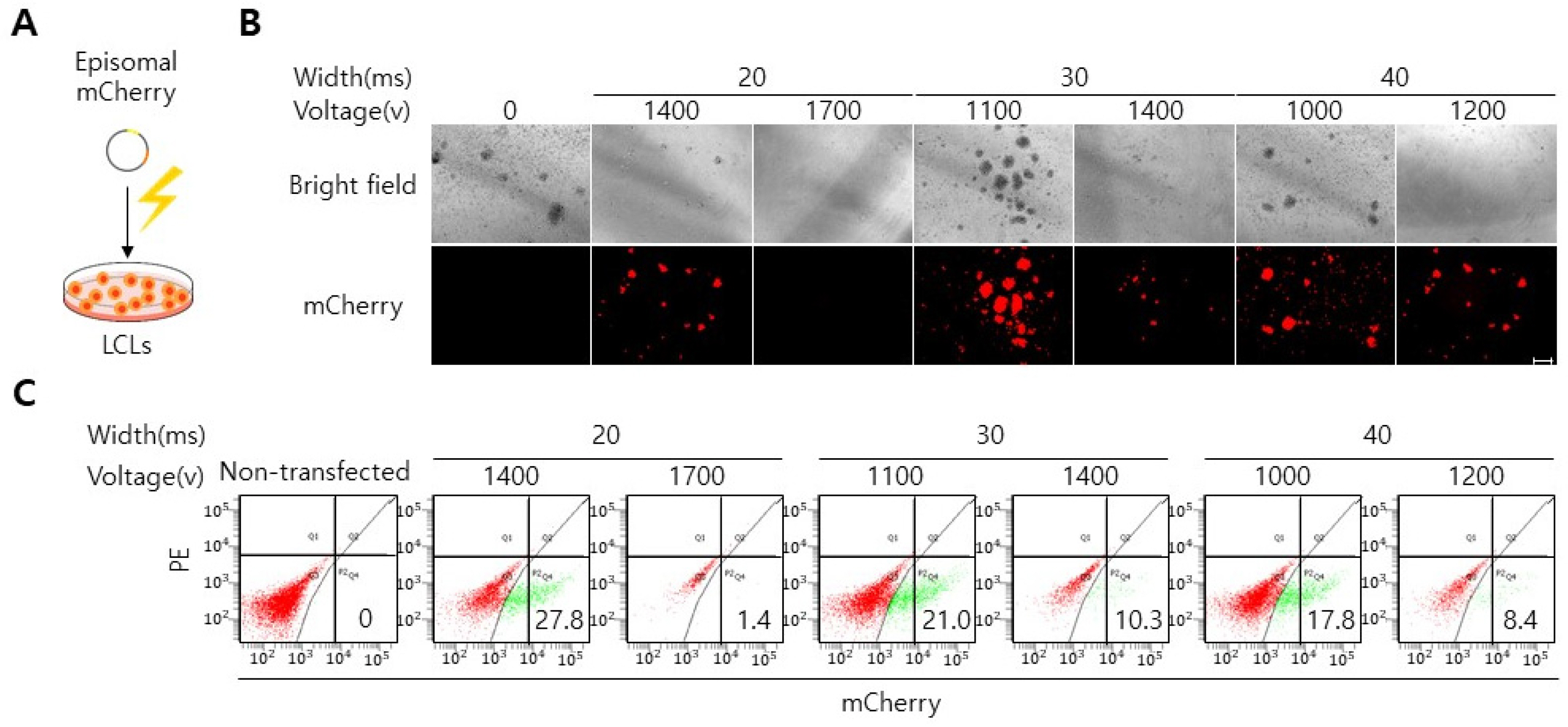
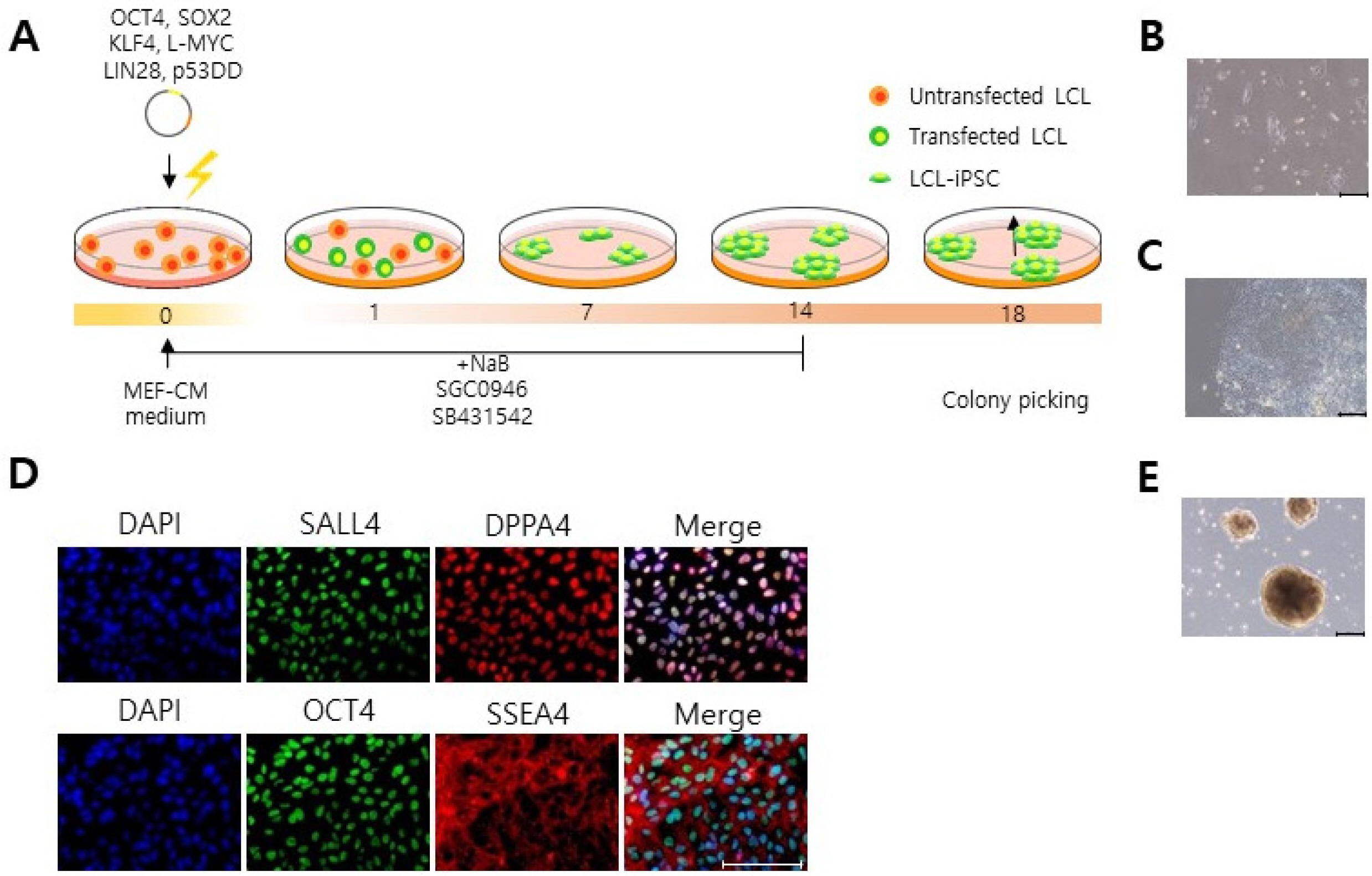
Here, we reveal an effective gene delivery method specifically for LCLs. We found that LCLs appear to be refractory toward retroviral and lentiviral transduction. Consequently, lentiviral and retroviral transduction of OCT4, SOX2, KFL4 and c-MYC into LCLs does not elicit iPSC colony formation. Interestingly, however we found that transfection of oriP/EBNA-1-based episomal vectors by electroporation is an efficient gene delivery system into LCLs, enabling iPSC generation from LCLs. These iPSCs expressed pluripotency makers (OCT4, NANOG, SSEA4, SALL4) and could form embryoid bodies.
Conclusions
Our data show that electroporation is an effective gene delivery method with which LCLs can be efficiently reprogrammed into iPSCs.
Figure
Reference
-
References
1. Takahashi K, Yamanaka S. 2016; A decade of transcription factor-mediated reprogramming to pluripotency. Nat Rev Mol Cell Biol. 17:183–193. DOI: 10.1038/nrm.2016.8. PMID: 26883003.2. Yamanaka S. 2020; Pluripotent stem cell-based cell therapy-promise and challenges. Cell Stem Cell. 27:523–531. DOI: 10.1016/j.stem.2020.09.014. PMID: 33007237.3. Inoue H, Nagata N, Kurokawa H, Yamanaka S. 2014; iPS cells: a game changer for future medicine. EMBO J. 33:409–417. DOI: 10.1002/embj.201387098. PMID: 24500035. PMCID: PMC3989624.4. Osório MJ, Goldman SA. 2018; Neurogenetics of Pelizaeus-Merzbacher disease. Handb Clin Neurol. 148:701–722. DOI: 10.1016/B978-0-444-64076-5.00045-4. PMID: 29478609. PMCID: PMC8167836.5. Elitt MS, Barbar L, Shick HE, Powers BE, Maeno-Hikichi Y, Madhavan M, Allan KC, Nawash BS, Gevorgyan AS, Hung S, Nevin ZS, Olsen HE, Hitomi M, Schlatzer DM, Zhao HT, Swayze A, LePage DF, Jiang W, Conlon RA, Rigo F, Tesar PJ. 2020; Suppression of proteolipid protein rescues Pelizaeus-Merzbacher disease. Nature. 585:397–403. DOI: 10.1038/s41586-020-2494-3. PMID: 32610343. PMCID: PMC7810164.6. Nevin ZS, Factor DC, Karl RT, Douvaras P, Laukka J, Windrem MS, Goldman SA, Fossati V, Hobson GM, Tesar PJ. 2017; Modeling the mutational and phenotypic landscapes of Pelizaeus-Merzbacher disease with human iPSC-derived oligodendrocytes. Am J Hum Genet. 100:617–634. DOI: 10.1016/j.ajhg.2017.03.005. PMID: 28366443. PMCID: PMC5384098.7. Nobuta H, Yang N, Ng YH, Marro SG, Sabeur K, Chavali M, Stockley JH, Killilea DW, Walter PB, Zhao C, Huie P Jr, Goldman SA, Kriegstein AR, Franklin RJM, Rowitch DH, Wernig M. 2019; Oligodendrocyte death in Pelizaeus-Merz-bacher disease is rescued by iron chelation. Cell Stem Cell. 25:531–541.e6. DOI: 10.1016/j.stem.2019.09.003. PMID: 31585094. PMCID: PMC8282124.8. Omi N, Tokuda Y, Ikeda Y, Ueno M, Mori K, Sotozono C, Kinoshita S, Nakano M, Tashiro K. 2017; Efficient and reliable establishment of lymphoblastoid cell lines by Epstein-Barr virus transformation from a limited amount of peripheral blood. Sci Rep. 7:43833. DOI: 10.1038/srep43833. PMID: 28272413. PMCID: PMC5341036.9. Lappalainen T, Sammeth M, Friedländer MR, Monlong J, Rivas MA, Gonzàlez-Porta M, Kurbatova N, Griebel T, Ferreira PG, Barann M, Wieland T, Greger L, van Iterson M, Almlöf J, Ribeca P, Pulyakhina I, Esser D, Giger T, Tikhonov A, Sultan M, Bertier G, MacArthur DG, Lek M, Lizano E, Buermans HP, Padioleau I, Schwarzmayr T, Karlberg O, Ongen H, Kilpinen H, Beltran S, Gut M, Kahlem K, Amstislavskiy V, Stegle O, Pirinen M, Mont-gomery SB, Donnelly P, McCarthy MI, Flicek P, Strom TM, Lehrach H, Schreiber S, Sudbrak R, Carracedo A, Antonarakis SE, Häsler R, Syvänen AC, van Ommen GJ, Brazma A, Meitinger T, Rosenstiel P, Guigó R, Gut IG, Estivill X, Dermitzakis ET. 't Hoen PA. Geuvadis Consortium. 2013; Transcriptome and genome sequencing uncovers functional variation in hu-mans. Nature. 501:506–511. DOI: 10.1038/nature12531. PMID: 24037378. PMCID: PMC3918453.10. Melé M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, Young TR, Goldmann JM, Pervouchine DD, Sullivan TJ, Johnson R, Segrè AV, Djebali S, Niarchou A, Wright FA, Lappalainen T, Calvo M, Getz G, Dermitzakis ET, Ardlie KG, Guigó R. GTEx Consortium. 2015; Human genomics. The human transcriptome across tissues and individuals. Science. 348:660–665. DOI: 10.1126/science.aaa0355. PMID: 25954002. PMCID: PMC4547472.11. ENCODE Project Consortium. 2012; An integrated encyclopedia of DNA elements in the human genome. Nature. 489:57–74. DOI: 10.1038/nature11247. PMID: 22955616. PMCID: PMC3439153.12. Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. 1000 Genomes Project Consortium. 2015; A global reference for human genetic variation. Nature. 526:68–74. DOI: 10.1038/nature15393. PMID: 26432245. PMCID: PMC4750478.13. Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Peltonen L, Dermitzakis E, Bonnen PE, Altshuler DM, Gibbs RA, de Bakker PI, Deloukas P, Gabriel SB, Gwilliam R, Hunt S, Inouye M, Jia X, Palotie A, Parkin M, Whittaker P, Yu F, Chang K, Hawes A, Lewis LR, Ren Y, Wheeler D, Gibbs RA, Muzny DM, Barnes C, Darvishi K, Hurles M, Korn JM, Kristiansson K, Lee C, McCarrol SA, Nemesh J, Dermitzakis E, Keinan A, Montgomery SB, Pollack S, Price AL, Soranzo N, Bonnen PE, Gibbs RA, Gonzaga-Jauregui C, Keinan A, Price AL, Yu F, Anttila V, Brodeur W, Daly MJ, Leslie S, McVean G, Moutsianas L, Nguyen H, Schaffner SF, Zhang Q, Ghori MJ, McGinnis R, McLaren W, Pollack S, Price AL, Schaffner SF, Takeuchi F, Grossman SR, Shlyakhter I, Hostetter EB, Sabeti PC, Adebamowo CA, Foster MW, Gordon DR, Licinio J, Manca MC, Marshall PA, Matsuda I, Ngare D, Wang VO, Reddy D, Rotimi CN, Royal CD, Sharp RR, Zeng C, Brooks LD, McEwen JE. International HapMap 3 Consortium. 2010; Integrating common and rare genetic variation in diverse human populations. Nature. 467:52–58. DOI: 10.1038/nature09298. PMID: 20811451. PMCID: PMC3173859.14. Kim KP, Li C, Bunina D, Jeong HW, Ghelman J, Yoon J, Shin B, Park H, Han DW, Zaugg JB, Kim J, Kuhlmann T, Adams RH, Noh KM, Goldman SA, Schöler HR. 2021; Donor cell memory confers a metastable state of directly converted cells. Cell Stem Cell. 28:1291–1306.e10. DOI: 10.1016/j.stem.2021.02.023. PMID: 33848472.15. Kim KP, Wu Y, Yoon J, Adachi K, Wu G, Velychko S, MacCarthy CM, Shin B, Röpke A, Arauzo-Bravo MJ, Stehling M, Han DW, Gao Y, Kim J, Gao S, Schöler HR. 2020; Reprogramming competence of OCT factors is determined by transactivation domains. Sci Adv. 6:eaaz7364. DOI: 10.1126/sciadv.aaz7364. PMID: 32917606. PMCID: PMC7467702.16. Warlich E, Kuehle J, Cantz T, Brugman MH, Maetzig T, Galla M, Filipczyk AA, Halle S, Klump H, Schöler HR, Baum C, Schroeder T, Schambach A. 2011; Lentiviral vector design and imaging approaches to visualize the early stages of cellular reprogramming. Mol Ther. 19:782–789. DOI: 10.1038/mt.2010.314. PMID: 21285961. PMCID: PMC3070104.17. Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012; Fiji: an open-source platform for biological-image analysis. Nat Methods. 9:676–682. DOI: 10.1038/nmeth.2019. PMID: 22743772. PMCID: PMC3855844.18. Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, Gonzalez F, Vassena R, Bilić J, Pekarik V, Tiscornia G, Edel M, Boué S, Izpisúa Belmonte JC. 2008; Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol. 26:1276–1284. DOI: 10.1038/nbt.1503. PMID: 18931654.19. Miller DG, Adam MA, Miller AD. 1990; Gene transfer by retrovirus vectors occurs only in cells that are actively replicating at the time of infection. Mol Cell Biol. 10:4239–4242. Erratum in: Mol Cell Biol 1992;12:433. DOI: 10.1128/MCB.10.8.4239. PMID: 2370865. PMCID: PMC360961.20. Kim KP, Choi J, Yoon J, Bruder JM, Shin B, Kim J, Arauzo-Bravo MJ, Han D, Wu G, Han DW, Kim J, Cramer P, Schöler HR. 2021; Permissive epigenomes endow reprogramming competence to transcriptional regulators. Nat Chem Biol. 17:47–56. DOI: 10.1038/s41589-020-0618-6. PMID: 32807969.21. Buchschacher GL Jr, Wong-Staal F. 2000; Development of lentiviral vectors for gene therapy for human diseases. Blood. 95:2499–2504. DOI: 10.1182/blood.V95.8.2499.008k35_2499_2504. PMID: 10753827.22. Harris E, Elmer JJ. 2021; Optimization of electroporation and other non-viral gene delivery strategies for T cells. Biotech-nol Prog. 37:e3066. DOI: 10.1002/btpr.3066. PMID: 32808434.23. Jordan ET, Collins M, Terefe J, Ugozzoli L, Rubio T. 2008; Optimizing electroporation conditions in primary and other difficult-to-transfect cells. J Biomol Tech. 19:328–334. PMID: 19183796. PMCID: PMC2628074.24. Hammerschmidt W, Sugden B. 2013; Replication of Epstein-Barr viral DNA. Cold Spring Harb Perspect Biol. 5:a013029. DOI: 10.1101/cshperspect.a013029. PMID: 23284049. PMCID: PMC3579399.25. Herbst F, Ball CR, Tuorto F, Nowrouzi A, Wang W, Zavidij O, Dieter SM, Fessler S, van der Hoeven F, Kloz U, Lyko F, Schmidt M, von Kalle C, Glimm H. 2012; Extensive methylation of promoter sequences silences lentiviral transgene expression during stem cell differentiation in vivo. Mol Ther. 20:1014–1021. DOI: 10.1038/mt.2012.46. PMID: 22434137. PMCID: PMC3345972.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Disease-specific pluripotent stem cells
- Generation and characterization of integration-free induced pluripotent stem cells from patients with autoimmune disease
- Comparison of Various Transfection Methods in Human and Bovine Cultured Cells
- Induced pluripotent stem cells and personalized medicine: current progress and future perspectives
- Generation of Induced Pluripotent Stem Cells and Neural Stem/Progenitor Cells from Newborns with Spina Bifida Aperta