Lab Med Online.
2022 Apr;12(2):116-121. 10.47429/lmo.2022.12.2.116.
Re-evaluation of the LDLR Gene Variants of Uncertain Significance Using ClinGen Guideline
- Affiliations
-
- 1Department of Laboratory Medicine and Genetics, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
- KMID: 2538594
- DOI: http://doi.org/10.47429/lmo.2022.12.2.116
Abstract
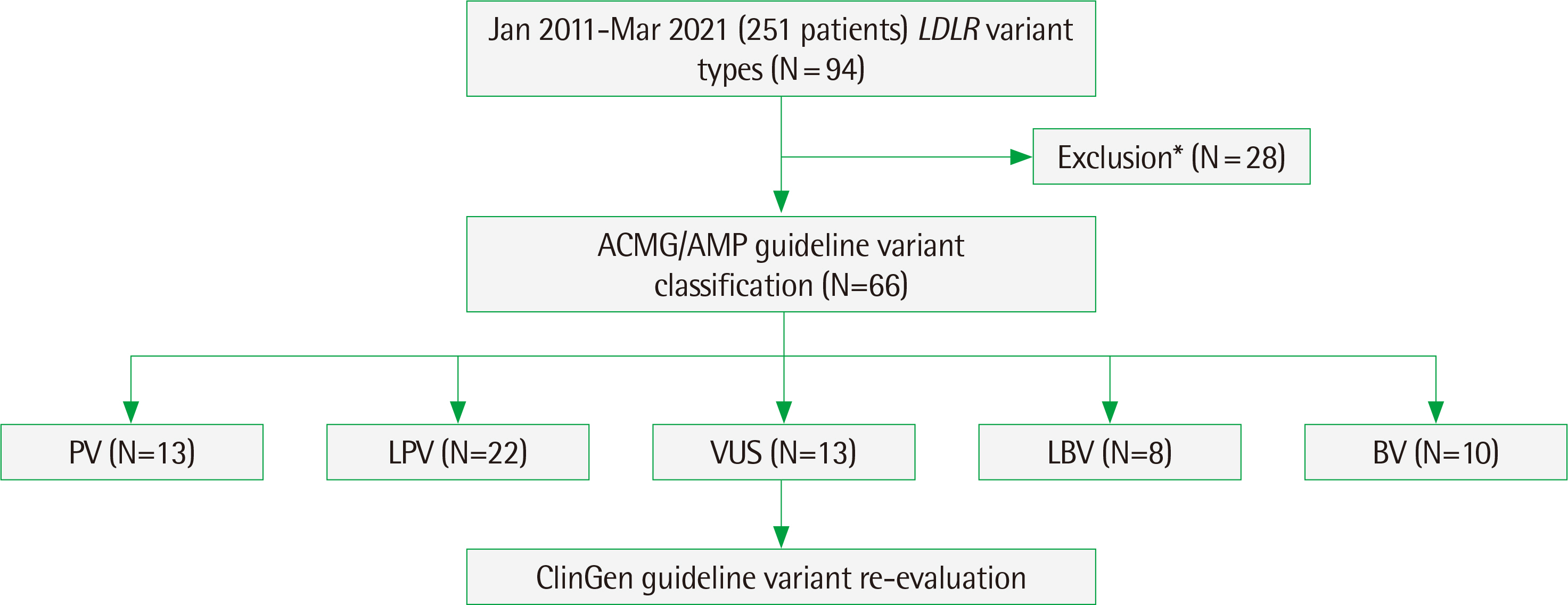
- Familial hypercholesterolemia (FH) is characterized by elevated LDL cholesterol levels, which is an important risk factor for early-onset cardiovascular disease. FH is one of the common genetic diseases that have a prevalence of approximately 1:250 and is associated with the heterozygous pathogenic variant of the gene encoding for the low-density lipoprotein receptor, LDLR gene (~90%). Variants identified through sequencing analysis are classified according to the American College of Medical Genetics and Association for Molecular Pathology Guideline. However, disease-specific variant curation is performed by the Clinical Genome Resource (ClinGen) Expert Panels. Since a specific ClinGen guideline for FH was recently published, here, we applied this new guideline to re-evaluate and re-classify what was reported as a Variant of Uncertain Significance (VUS) in the LDLR gene. Among 66 different types of LDLR variants from 251 patients during 2011 and 2021, 13 different VUSs reported from 15 patients were re-classified. Among 13 VUSs, two missense variants; c.268G>T (p.D90Y) and c.694G>T (p.A232S), were re-classified to likely pathogenic variants. Using the ClinGen guideline, VUSs previously lacking sufficient evidence to be classified as likely pathogenic have now been appropriately classified. Considering the genetic causes and prevalence of FH, the evaluation and classification of the LDLR gene variants should be updated according to the new ClinGen guideline specific for FH.
Keyword
Figure
Reference
-
1. Akioyamen LE, Genest J, Shan SD, Reel RL, Albaum JM, Chu A, et al. 2017; Estimating the prevalence of heterozygous familial hypercholesterolaemia: a systematic review and meta-analysis. BMJ Open. 7:e016461. DOI: 10.1136/bmjopen-2017-016461. PMID: 28864697. PMCID: PMC5588988.
Article2. Rhee EJ, Kim HC, Kim JH, Lee EY, Kim BJ, Kim EM, et al. 2019; 2018 Guidelines for the management of dyslipidemia. Korean J Intern Med. 34:723–71. DOI: 10.3904/kjim.2019.188. PMID: 31272142. PMCID: PMC6610190.
Article3. Nordestgaard BG, Chapman MJ, Humphries SE, Ginsberg HN, Masana L, Descamps OS, et al. 2013; Familial hypercholesterolaemia is underdiagnosed and undertreated in the general population: guidance for clinicians to prevent coronary heart disease: consensus statement of the European Atherosclerosis Society. Eur Heart J. 34:3478–90. DOI: 10.1093/eurheartj/eht273. PMID: 23956253. PMCID: PMC3844152.4. Nordestgaard BG, Benn M. 2017; Genetic testing for familial hypercholesterolaemia is essential in individuals with high LDL cholesterol: who does it in the world? Eur Heart J. 38:1580–3. DOI: 10.1093/eurheartj/ehx136. PMID: 28419271.
Article5. Soutar AK, Naoumova RP. 2007; Mechanisms of disease: genetic causes of familial hypercholesterolemia. Nat Clin Pract Cardiovasc Med. 4:214–25. DOI: 10.1038/ncpcardio0836. PMID: 17380167.
Article6. Defesche JC, Lansberg PJ, Umans-Eckenhausen MA, Kastelein JJ. 2004; Advanced method for the identification of patients with inherited hypercholesterolemia. Semin Vasc Med. 4:59–65. DOI: 10.1055/s-2004-822987. PMID: 15199434.
Article7. Risk of fatal coronary heart disease in familial hypercholesterolaemia. 1991; Scientific Steering Committee on behalf of the Simon Broome Register Group. BMJ. 303:893–6. DOI: 10.1136/bmj.303.6807.893. PMID: 1933004. PMCID: PMC1671226.8. Dedoussis GV, Schmidt H, Genschel J. 2004; LDL-receptor mutations in Europe. Hum Mutat. 24:443–59. DOI: 10.1002/humu.20105. PMID: 15523646.
Article9. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. 2015; Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 17:405–24. DOI: 10.1038/gim.2015.30. PMID: 25741868. PMCID: PMC4544753.
Article10. Rehm HL, Berg JS, Brooks LD, Bustamante CD, Evans JP, Landrum MJ, et al. 2015; ClinGen--the Clinical Genome Resource. N Engl J Med. 372:2235–42. DOI: 10.1056/NEJMsr1406261. PMID: 26014595. PMCID: PMC4474187.11. Chora JR, Medeiros AM, Alves AC, Bourbon M. 2018; Analysis of publicly available LDLR, APOB, and PCSK9 variants associated with familial hypercholesterolemia: application of ACMG guidelines and implications for familial hypercholesterolemia diagnosis. Genet Med. 20:591–8. DOI: 10.1038/gim.2017.151. PMID: 29261184.12. Clinical Genome Resource. ClinGen Familial Hypercholesterolemia Expert Panel Specifications to the ACMG/AMP Variant Interpretation Guidelines Version 1. https://clinicalgenome.org/site/assets/files/6301/clingen_fh_acmg_specifications_v1_1-1.pdf. Updated on Apr 2021.13. Vears DF, Niemiec E, Howard HC, Borry P. 2018; Analysis of VUS reporting, variant reinterpretation and recontact policies in clinical genomic sequencing consent forms. Eur J Hum Genet. 26:1743–51. DOI: 10.1038/s41431-018-0239-7. PMID: 30143804. PMCID: PMC6244391.
Article14. den Dunnen JT, Dalgleish R, Maglott DR, Hart RK, Greenblatt MS, McGowan-Jordan J, et al. 2016; HGVS Recommendations for the Description of Sequence Variants: 2016 Update. Hum Mutat. 37:564–9. DOI: 10.1002/humu.22981. PMID: 26931183.
Article15. Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, et al. 2016; REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am J Hum Genet. 99:877–85. DOI: 10.1016/j.ajhg.2016.08.016. PMID: 27666373. PMCID: PMC5065685.
Article16. Yeo G, Burge CB. 2004; Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 11:377–94. DOI: 10.1089/1066527041410418. PMID: 15285897.
Article17. Iacocca MA, Chora JR, Carrié A, Freiberger T, Leigh SE, Defesche JC, et al. 2018; ClinVar database of global familial hypercholesterolemia-associated DNA variants. Hum Mutat. 39:1631–40. DOI: 10.1002/humu.23634. PMID: 30311388. PMCID: PMC6206854.
Article18. Deignan JL, Chung WK, Kearney HM, Monaghan KG, Rehder CW, Chao EC. 2019; Points to consider in the reevaluation and reanalysis of genomic test results: a statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 21:1267–70. DOI: 10.1038/s41436-019-0478-1. PMID: 31015575. PMCID: PMC6559819.
Article19. Association for Clinical Genomic Science. 2020. ACGS Best Practice Guidelines for Variant Classification in Rare Disease 2020. https://www.acgs.uk.com/media/11631/uk-practice-guidelines-for-variant-classification-v4-01-2020.pdf. Updated on Feb 2020.20. Rodríguez-Jiménez C, Pernía O, Mostaza J, Rodríguez-Antolín C, de Dios García-Díaz J, Alonso-Cerezo C, et al. 2019; Functional analysis of new variants at the low-density lipoprotein receptor associated with familial hypercholesterolemia. Hum Mutat. 40:1181–90. DOI: 10.1002/humu.23801. PMID: 31106925.21. Huang CC, Charng MJ. 2020; Genetic diagnosis of familial hypercholesterolemia in Asia. Front Genet. 11:833. DOI: 10.3389/fgene.2020.00833. PMID: 32793292. PMCID: PMC7393677.
Article22. Etxebarria A, Benito-Vicente A, Stef M, Ostolaza H, Palacios L, Martin C. 2015; Activity-associated effect of LDL receptor missense variants located in the cysteine-rich repeats. Atherosclerosis. 238:304–12. DOI: 10.1016/j.atherosclerosis.2014.12.026. PMID: 25545329.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Re-evaluation of a Fibrillin-1 Gene Variant of Uncertain Significance Using the ClinGen Guidelines
- Detection of Familial Hypercholesterolemia Using Next Generation Sequencing in Two Population-Based Cohorts
- Association of Clinical Characteristics With Familial Hypercholesterolaemia Variants in a Lipid Clinic Setting: A Case-Control Study
- Sterol-independent repression of low density lipoprotein receptor promoter by peroxisome proliferator activated receptor gamma coactivator-1alpha (PGC-1alpha)
- Method-Based Proficiency Test Program for Assessing Quality of Sanger Sequencing-Based Molecular Tests