Cancer Res Treat.
2023 Jan;55(1):291-303. 10.4143/crt.2022.017.
Circulating Tumor DNA–Based Genotyping and Monitoring for Predicting Disease Relapses of Patients with Peripheral T-Cell Lymphomas
- Affiliations
-
- 1Division of Hematology-Oncology, Department of Medicine, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
- 2Department of Health Sciences and Technology, Samsung Advanced Institute for Health Sciences and Technology, Sungkyunkwan University School of Medicine, Seoul, Korea
- 3Samsung Genome Institute Samsung Medical Center, Seoul, Korea
- 4GENINUS Inc., Seoul, Korea
- 5Department of Laboratory Medicine and Genetics, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
- 6Department of Pathology, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
- KMID: 2538012
- DOI: http://doi.org/10.4143/crt.2022.017
Abstract
- Purpose
Plasma circulating tumor DNA (ctDNA) could reflect the genetic alterations present in tumor tissues. However, there is little information about the clinical relevance of cell-free DNA genotyping in peripheral T-cell lymphoma (PTCL).
Materials and Methods
After targeted sequencing plasma cell-free DNA of patients with various subtypes of PTCL (n=94), we analyzed the mutation profiles of plasma ctDNA samples and their predictive value of dynamic ctDNA monitoring for treatment outcomes.
Results
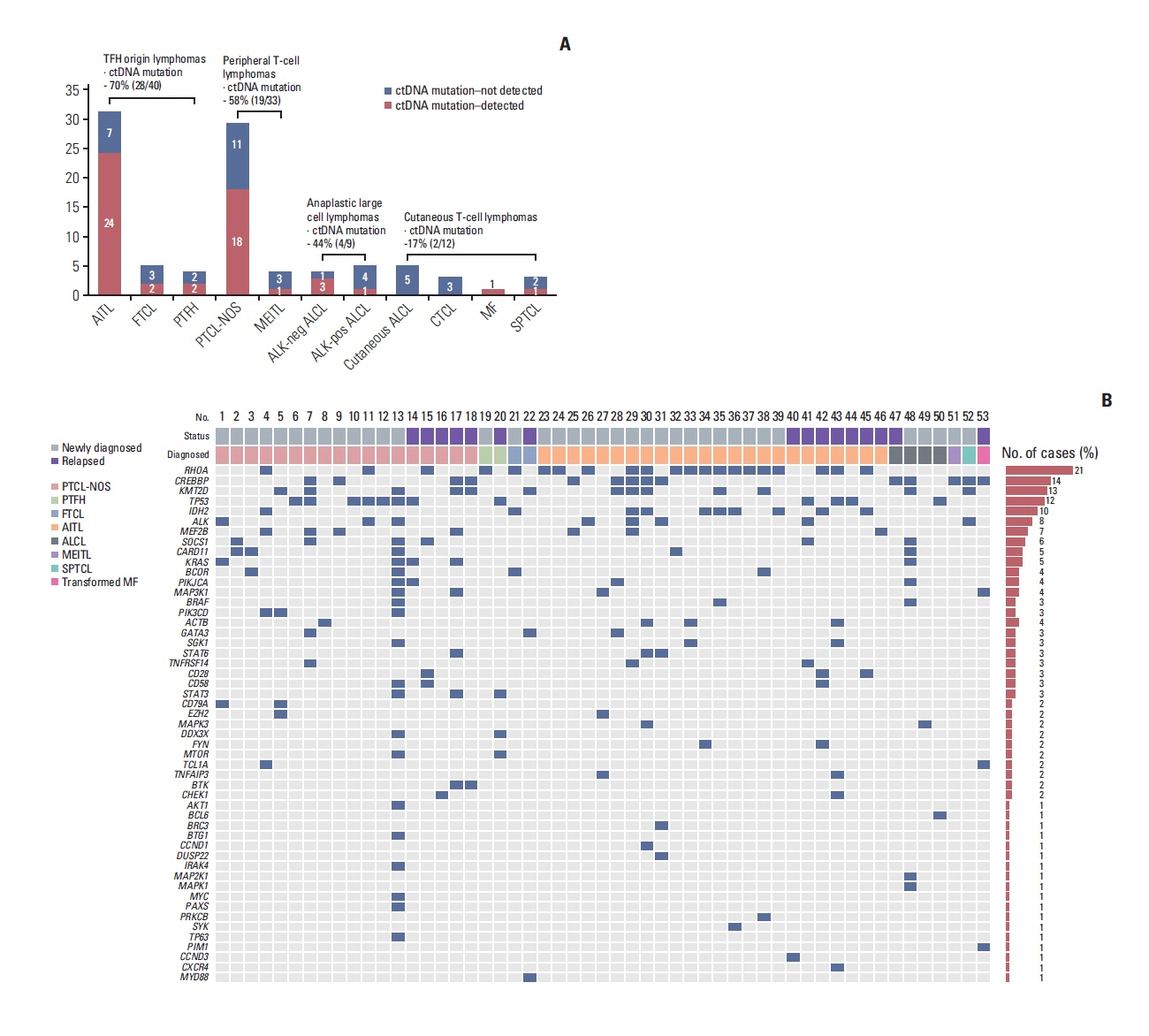
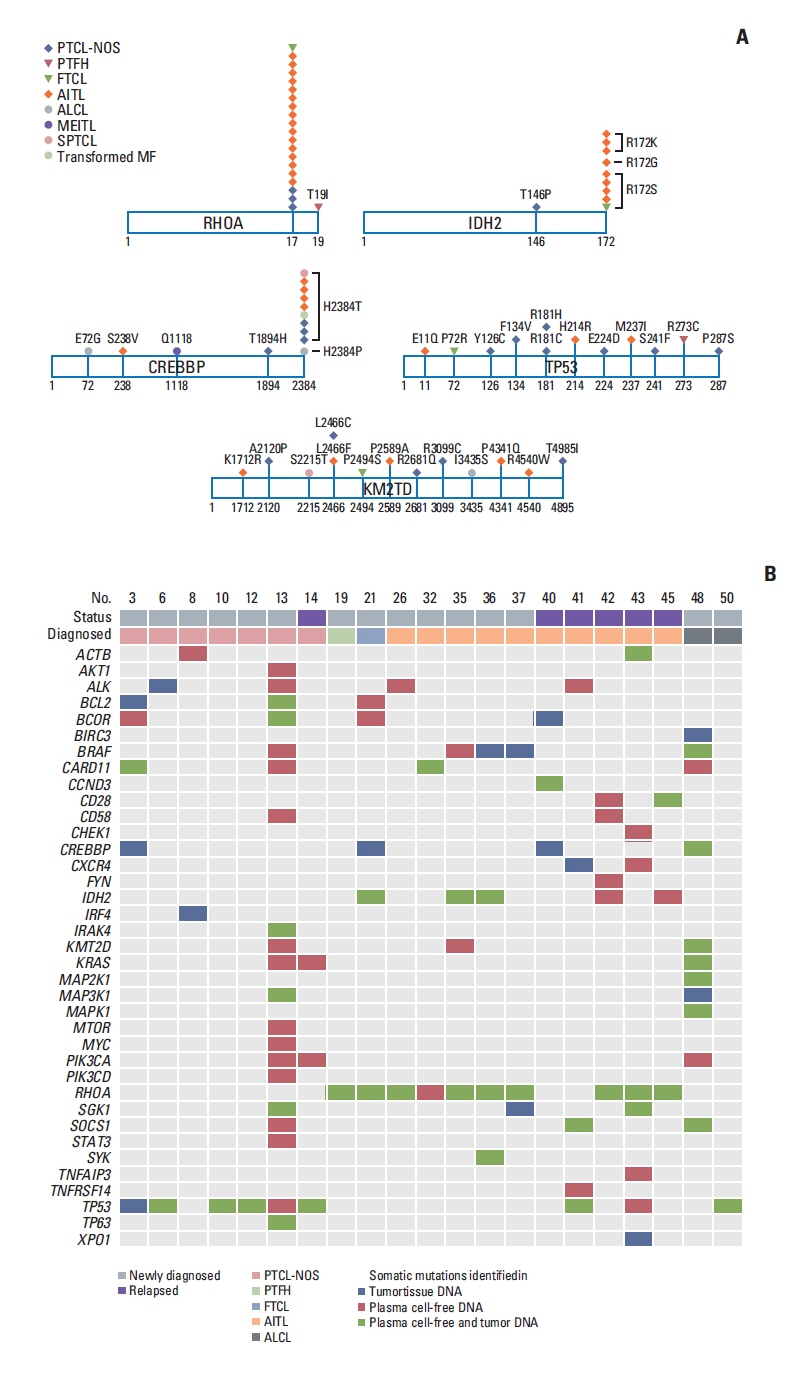
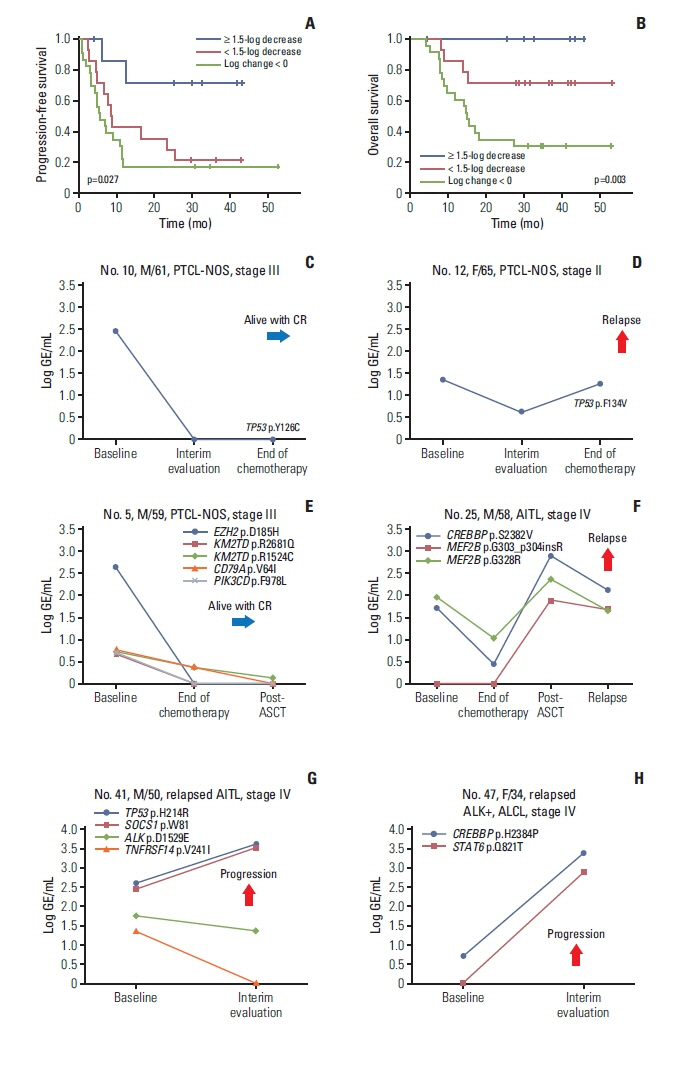
Plasma ctDNA mutations were detected in 53 patients (56%, 53/94), and the detection rate of somatic mutations was highest in angioimmunoblastic T-cell lymphoma (24/31, 77%) and PTCL, not otherwise specified (18/29, 62.1%). Somatic mutations were detected in 51 of 66 genes that were sequenced, including the following top 10 ranked genes: RHOA, CREBBP, KMT2D, TP53, IDH2, ALK, MEF2B, SOCS1, CARD11, and KRAS. In the longitudinal assessment of ctDNA mutation, the difference in ctDNA mutation volume after treatment showed a significant correlation with disease relapse or progression. Thus, a ≥ 1.5-log decrease in genome equivalent (GE) between baseline and the end of treatment showed a significant association with better survival outcomes than a < 1.5-log decrease in GE.
Conclusion
Our results suggest the clinical relevance of plasma ctDNA analysis in patients with PTCL. However, our findings should be validated by a subsequent study with a larger study population and using a broader gene panel.
Figure
Cited by 1 articles
-
Feasibility of Circulating Tumor DNA Analysis in Patients with Follicular Lymphoma
Sang Eun Yoon, Seung-Ho Shin, Dae Keun Nam, Junhun Cho, Won Seog Kim, Seok Jin Kim
Cancer Res Treat. 2024;56(3):920-935. doi: 10.4143/crt.2023.869.
Reference
-
References
1. Vose J, Armitage J, Weisenburger D. International T-Cell Lymphoma Project. International peripheral T-cell and natural killer/T-cell lymphoma study: pathology findings and clinical outcomes. J Clin Oncol. 2008; 26:4124–30.2. Briski R, Feldman AL, Bailey NG, Lim MS, Ristow K, Habermann TM, et al. The role of front-line anthracycline-containing chemotherapy regimens in peripheral T-cell lymphomas. Blood Cancer J. 2014; 4:e214.3. Ellin F, Landstrom J, Jerkeman M, Relander T. Real-world data on prognostic factors and treatment in peripheral T-cell lymphomas: a study from the Swedish Lymphoma Registry. Blood. 2014; 124:1570–7.4. Phan A, Veldman R, Lechowicz MJ. T-cell lymphoma epidemiology: the known and unknown. Curr Hematol Malig Rep. 2016; 11:492–503.5. Ma H, Marchi E, O’Connor OA. The peripheral T-cell lymphomas: an unusual path to cure. Lancet Haematol. 2020; 7:e765–71.
Article6. Cirillo M, Craig AFM, Borchmann S, Kurtz DM. Liquid biopsy in lymphoma: molecular methods and clinical applications. Cancer Treat Rev. 2020; 91:102106.7. Dogliotti I, Drandi D, Genuardi E, Ferrero S. New molecular technologies for minimal residual disease evaluation in B-cell lymphoid malignancies. J Clin Med. 2018; 7:288.8. Camus V, Jardin F. Cell-free DNA and the monitoring of lymphoma treatment. Pharmacogenomics. 2019; 20:1271–82.
Article9. Kurtz DM, Scherer F, Jin MC, Soo J, Craig AF, Esfahani MS, et al. Circulating tumor DNA measurements as early outcome predictors in diffuse large B-cell lymphoma. J Clin Oncol. 2018; 36:2845–53.10. Qi F, Cao Z, Chen B, Chai Y, Lin J, Ye J, et al. Liquid biopsy in extranodal NK/T-cell lymphoma: a prospective analysis of cell-free DNA genotyping and monitoring. Blood Adv. 2021; 5:2505–14.
Article11. Miljkovic MD, Melani C, Pittaluga S, Lakhotia R, Lucas N, Jacob A, et al. Next-generation sequencing-based monitoring of circulating tumor DNA reveals clonotypic heterogeneity in untreated PTCL. Blood Adv. 2021; 5:4198–210.12. Zhang W, Wang W, Han X, Gan Y, Qian L, Zhang Y, et al. Circulating tumor DNA by high-throughput sequencing of T cell receptor monitored treatment response and predicted treatment failure in T cell lymphomas. Int J Lab Hematol. 2021; 43:1041–9.
Article13. Sakata-Yanagimoto M, Nakamoto-Matsubara R, Komori D, Nguyen TB, Hattori K, Nanmoku T, et al. Detection of the circulating tumor DNAs in angioimmunoblastic T-cell lymphoma. Ann Hematol. 2017; 96:1471–5.14. Shin SH, Kim YJ, Lee D, Cho D, Ko YH, Cho J, et al. Analysis of circulating tumor DNA by targeted ultra-deep sequencing across various non-Hodgkin lymphoma subtypes. Leuk Lymphoma. 2019; 60:2237–46.15. Hur JY, Kim YJ, Yoon SE, Son DS, Park WY, Kim SJ, et al. Plasma cell-free DNA is a prognostic biomarker for survival in patients with aggressive non-Hodgkin lymphomas. Ann Hematol. 2020; 99:1293–302.16. Cheson BD, Fisher RI, Barrington SF, Cavalli F, Schwartz LH, Zucca E, et al. Recommendations for initial evaluation, staging, and response assessment of Hodgkin and non-Hodgkin lymphoma: the Lugano classification. J Clin Oncol. 2014; 32:3059–68.17. Newman AM, Lovejoy AF, Klass DM, Kurtz DM, Chabon JJ, Scherer F, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol. 2016; 34:547–55.18. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010; 20:1297–303.19. Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011; 27:2987–93.20. Avanzini S, Kurtz DM, Chabon JJ, Moding EJ, Hori SS, Gambhir SS, et al. A mathematical model of ctDNA shedding predicts tumor detection size. Sci Adv. 2020; 6:eabc4308.21. Sakata-Yanagimoto M, Enami T, Yoshida K, Shiraishi Y, Ishii R, Miyake Y, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet. 2014; 46:171–5.22. Yoo HY, Sung MK, Lee SH, Kim S, Lee H, Park S, et al. A recurrent inactivating mutation in RHOA GTPase in angioimmunoblastic T cell lymphoma. Nat Genet. 2014; 46:371–5.23. Palomero T, Couronne L, Khiabanian H, Kim MY, Ambesi-Impiombato A, Perez-Garcia A, et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat Genet. 2014; 46:166–70.24. Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016; 127:2375–90.25. Lemonnier F, Safar V, Beldi-Ferchiou A, Cottereau AS, Bachy E, Cartron G, et al. Integrative analysis of a phase 2 trial combining lenalidomide with CHOP in angioimmunoblastic T-cell lymphoma. Blood Adv. 2021; 5:539–48.26. Heavican TB, Bouska A, Yu J, Lone W, Amador C, Gong Q, et al. Genetic drivers of oncogenic pathways in molecular subgroups of peripheral T-cell lymphoma. Blood. 2019; 133:1664–76.27. Ji MM, Huang YH, Huang JY, Wang ZF, Fu D, Liu H, et al. Histone modifier gene mutations in peripheral T-cell lymphoma not otherwise specified. Haematologica. 2018; 103:679–87.28. Xie C, Li X, Zeng H, Qian W. Molecular insights into pathogenesis and targeted therapy of peripheral T cell lymphoma. Exp Hematol Oncol. 2020; 9:30.29. Melchardt T, Hufnagl C, Weinstock DM, Kopp N, Neureiter D, Trankenschuh W, et al. Clonal evolution in relapsed and refractory diffuse large B-cell lymphoma is characterized by high dynamics of subclones. Oncotarget. 2016; 7:51494–502.
Article30. Pon JR, Wong J, Saberi S, Alder O, Moksa M, Grace Cheng SW, et al. MEF2B mutations in non-Hodgkin lymphoma dysregulate cell migration by decreasing MEF2B target gene activation. Nat Commun. 2015; 6:7953.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Clinical Application of Circulating Tumor DNA Analysis
- Current Methods of Circulating Tumor Cell Detection
- Circulating Tumor Cells and Extracellular Nucleic Acids in Breast Cancer
- Circulating Cell-free Tumor Nucleic Acids in Gastric Cancer
- Circulating Lymphoma Cells in the Peripheral Blood from 4 Cases of Mantle and T Cell Types of Non-Hodgkin's Lymphoma: Light and Electron Microscopic Morphology