Cardiovasc Prev Pharmacother.
2021 Jul;3(3):64-72. 10.36011/cpp.2021.3.e9.
Perceptron: Basic Principles of Deep Neural Networks
- Affiliations
-
- 1Department of Artificial Intelligence and Software Technology, Sunmoon University, Asan, Korea
- 2Department of Medical Informatics, College of Medicine, The Catholic University of Korea, Seoul, Korea
- 3Division of Endocrinology and Metabolism, Department of Internal Medicine, Seoul St. Mary's Hospital, College of Medicine, The Catholic University of Korea, Seoul, Korea
- KMID: 2536942
- DOI: http://doi.org/10.36011/cpp.2021.3.e9
Abstract
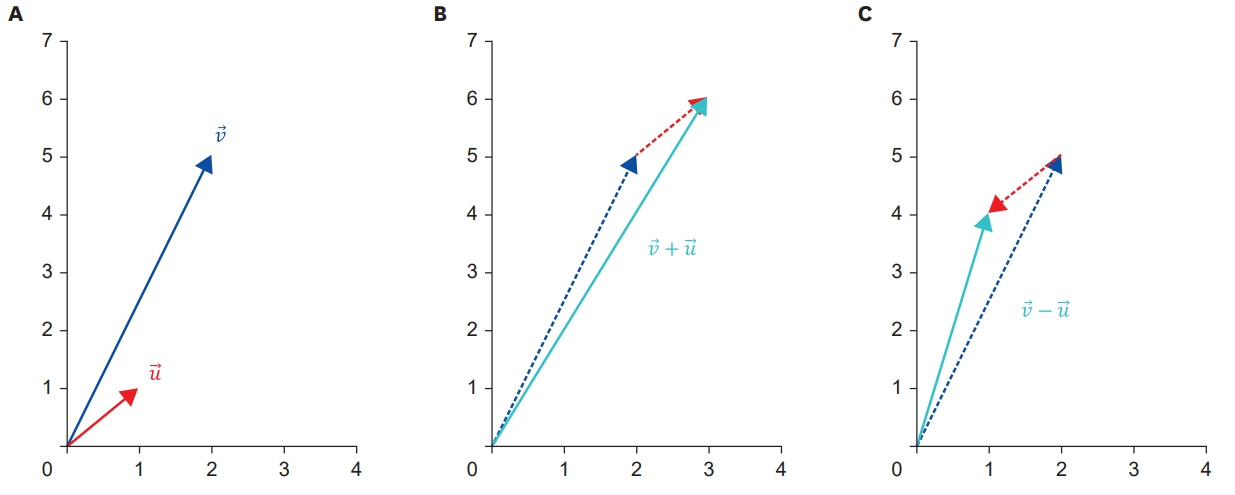
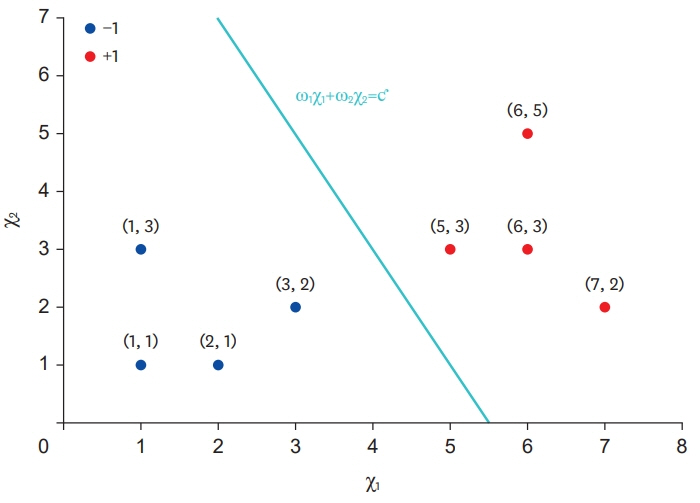
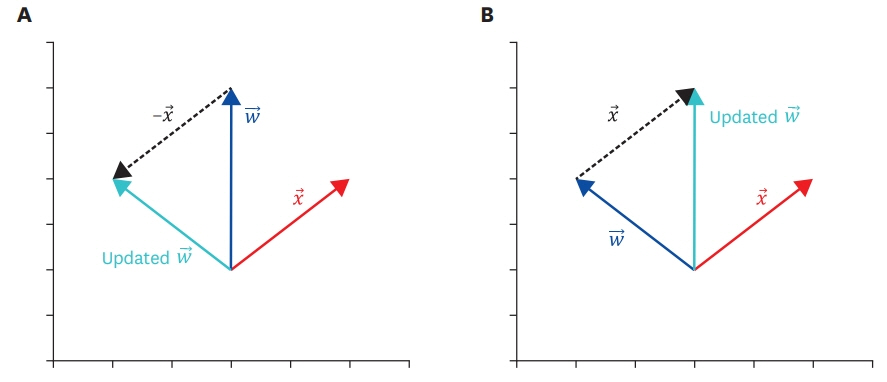
- Big data, artificial intelligence, machine learning, and deep learning have received considerable attention in the medical field. Attempts to use such machine learning in areas where medical decisions are difficult or necessary are continuously being made. To date, there have been many attempts to solve problems associated with the use of machine learning by using deep learning; hence, physicians should also have basic knowledge in this regard. Deep neural networks are one of the most actively studied methods in the field of machine learning. The perceptron is one of these artificial neural network models, and it can be considered as the starting point of artificial neural network models. Perceptrons receive various inputs and produce one output. In a perceptron, various weights (ω) are given to various inputs, and as ω becomes larger, it becomes an important factor. In other words, a perceptron is an algorithm with both input and output. When an input is provided, the output is produced according to a set rule. In this paper, the decision rules of the perceptron and its basic principles are examined. The intent is to provide a wide range of physicians with an understanding of the latest machine-learning methodologies based on deep neural networks.
Figure
Reference
-
1. Mitchell TM. Machine Learning. 1st ed. New York, NY: McGraw-Hill;1997.2. Kim HS, Yoon KH. Lessons from use of continuous glucose monitoring systems in digital healthcare. Endocrinol Metab. 2020; 35:541–8.
Article3. Lee H, Kim HS. Logistic regression and least absolute shrinkage and selection operator. Cardiovasc Prev Pharmacother. 2020; 2:142–6.
Article4. Lee M, Kim H, Joe H, Kim HG. Multi-channel PINN: investigating scalable and transferable neural networks for drug discovery. J Cheminform. 2019; 11:46.
Article5. Miotto R, Li L, Kidd BA, Dudley JT. Deep patient: an unsupervised representation to predict the future of patients from the electronic health records. Sci Rep. 2016; 6:26094.
Article6. Senior AW, Evans R, Jumper J, Kirkpatrick J, Sifre L, Green T, Qin C, Žídek A, Nelson AW, Bridgland A, Penedones H, Petersen S, Simonyan K, Crossan S, Kohli P, Jones DT, Silver D, Kavukcuoglu K, Hassabis D. Improved protein structure prediction using potentials from deep learning. Nature. 2020; 577:706–10.
Article7. Xu J, Zhang H, Zheng J, Dovoedo P, Yin Y. eCAMI: simultaneous classification and motif identification for enzyme annotation. Bioinformatics. 2020; 36:2068–75.
Article8. Russell S, Norvind P. Artificial Intelligence: a Modern Approach. 3rd ed. Englewood Cliffs, NJ: Prentice Hall;2010.9. Herold D, Lutter D, Schachtner R, Tome AM, Schmitz G, Lang EW. Comparison of unsupervised and supervised gene selection methods. Annu Int Conf IEEE Eng Med Biol Soc. 2008; 2008:5212–5.
Article10. Goudbeek M, Swingley D, Smits R. Supervised and unsupervised learning of multidimensional acoustic categories. J Exp Psychol Hum Percept Perform. 2009; 35:1913–33.
Article11. Dreiseitl S, Ohno-Machado L. Logistic regression and artificial neural network classification models: a methodology review. J Biomed Inform. 2002; 35:352–9.
Article12. Lorencin I; Anđelić N, Španjol J, Car Z. Using multi-layer perceptron with Laplacian edge detector for bladder cancer diagnosis. Artif Intell Med. 2020; 102:101746.
Article13. Castro W, Oblitas J, Santa-Cruz R, Avila-George H. Multilayer perceptron architecture optimization using parallel computing techniques. PLoS One. 2017; 12:e0189369.
Article14. Metsker O, Magoev K, Yanishevskiy S, Yakovlev A, Kopanitsa G, Zvartau N. Identification of diabetes risk factors in chronic cardiovascular patients. Stud Health Technol Inform. 2020; 273:136–41.15. Rossi F, Conan-Guez B. Theoretical properties of projection based multilayer perceptrons with functional inputs. Neural Process Lett. 2006; 23:55–70.
Article16. Ghosh K, Stuke A, Todorović M; Jørgensen PB, Schmidt MN, Vehtari A, Rinke P. Deep learning spectroscopy: neural networks for molecular excitation spectra. Adv Sci (Weinh). 2019; 6:1801367.
Article17. Shafi N, Bukhari F, Iqbal W, Almustafa KM, Asif M, Nawaz Z. Cleft prediction before birth using deep neural network. Health Informatics J. 2020; 26:2568–85.
Article18. Kim EH, Oh SK, Pedrycz W. Design of double fuzzy clustering-driven context neural networks. Neural Netw. 2018; 104:1–14.
Article19. Li Y, Zhang T. Deep neural mapping support vector machines. Neural Netw. 2017; 93:185–94.
Article20. Hong S, Zhou Y, Wu M, Shang J, Wang Q, Li H, Xie J. Combining deep neural networks and engineered features for cardiac arrhythmia detection from ECG recordings. Physiol Meas. 2019; 40:054009.
Article21. Acharya UR, Oh SL, Hagiwara Y, Tan JH, Adam M, Gertych A, Tan RS. A deep convolutional neural network model to classify heartbeats. Comput Biol Med. 2017; 89:389–96.
Article22. Cai L, Gao J, Zhao D. A review of the application of deep learning in medical image classification and segmentation. Ann Transl Med. 2020; 8:713.
Article23. Mishra S, Banerjee M. Automatic caption generation of retinal diseases with self-trained RNN merge model. In : Chaki R, Cortesi A, Saeed K, Chaki N, editors. Advanced Computing and Systems for Security. Advances in Intelligent Systems and Computing, Vol. 1136. Springer: Singapore;2020. p. 1–10.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Prediction of oculocardiac reflex in strabismus surgery using neural networks
- On the Use of Neural Networks for the Risk Factor Analysis of NIDDM
- Development of Breast Cancer Predication Model Using Neural Networks
- Anesthesia research in the artificial intelligence era
- Artificial Neural Network: Understanding the Basic Concepts without Mathematics