Clin Exp Otorhinolaryngol.
2022 Nov;15(4):372-379. 10.21053/ceo.2022.00150.
The Expression of Defensin-Associated Genes May Be Correlated With Lymph Node Metastasis of Early-Stage Tongue Cancer
- Affiliations
-
- 1Department of Otorhinolaryngology-Head and Neck Surgery, Seoul National University Boramae Medical Center, Seoul, Korea
- 2Department of Otorhinolaryngology-Head and Neck Surgery, THANQ Seoul Thyroid-Head and Neck Surgery Center, Seoul, Korea
- 3Department of Otorhinolaryngology-Head and Neck Surgery, Seoul National University Bundang Hospital, Seongnam, Korea
- 4Department of Otorhinolaryngology-Head and Neck Surgery, Seoul National University Hospital, Seoul, Korea
- 5Sensory Organ Research Institute, Seoul National University Medical Research Center, Seoul, Korea
- 6Cancer Research Institute, Seoul National University, Seoul, Korea
- KMID: 2536536
- DOI: http://doi.org/10.21053/ceo.2022.00150
Abstract
Objectives
. We aimed to assess the genetic differences between cases of early-stage tongue cancer that were positive or negative for lymph node metastasis.
Methods
. In total, 35 cases of tongue cancer with RNA sequencing data were enrolled in this study. The gene expression profile of the following two groups was compared: N0 group (T stage 1 or 2 with N0 stage) and N+ group (T stage 1 or 2 with N+ stage). Using the R and limma packages in the Bioconductor program, we extracted the differentially expressed genes (DEGs). Gene ontology and pathway enrichment analysis were performed using the Database for Annotation, Visualization and Integration Discovery (DAVID) online tool. Immune cell infiltration was analyzed using the CIBERSORT online program. Immunochemical staining of the cancer tissue was evaluated and The Cancer Genome Atlas (TCGA) data were analyzed to validate the identified DEGs.
Results
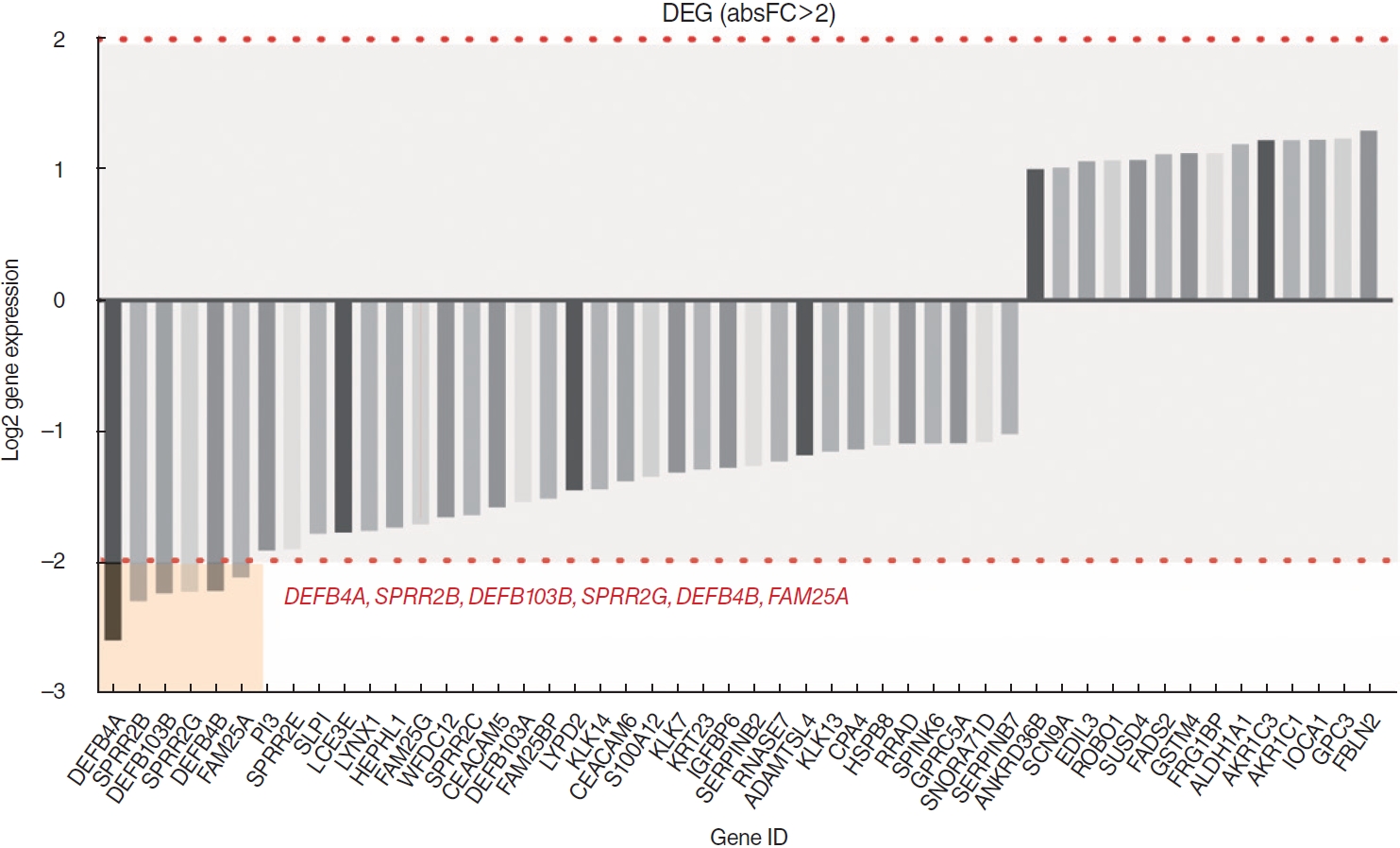
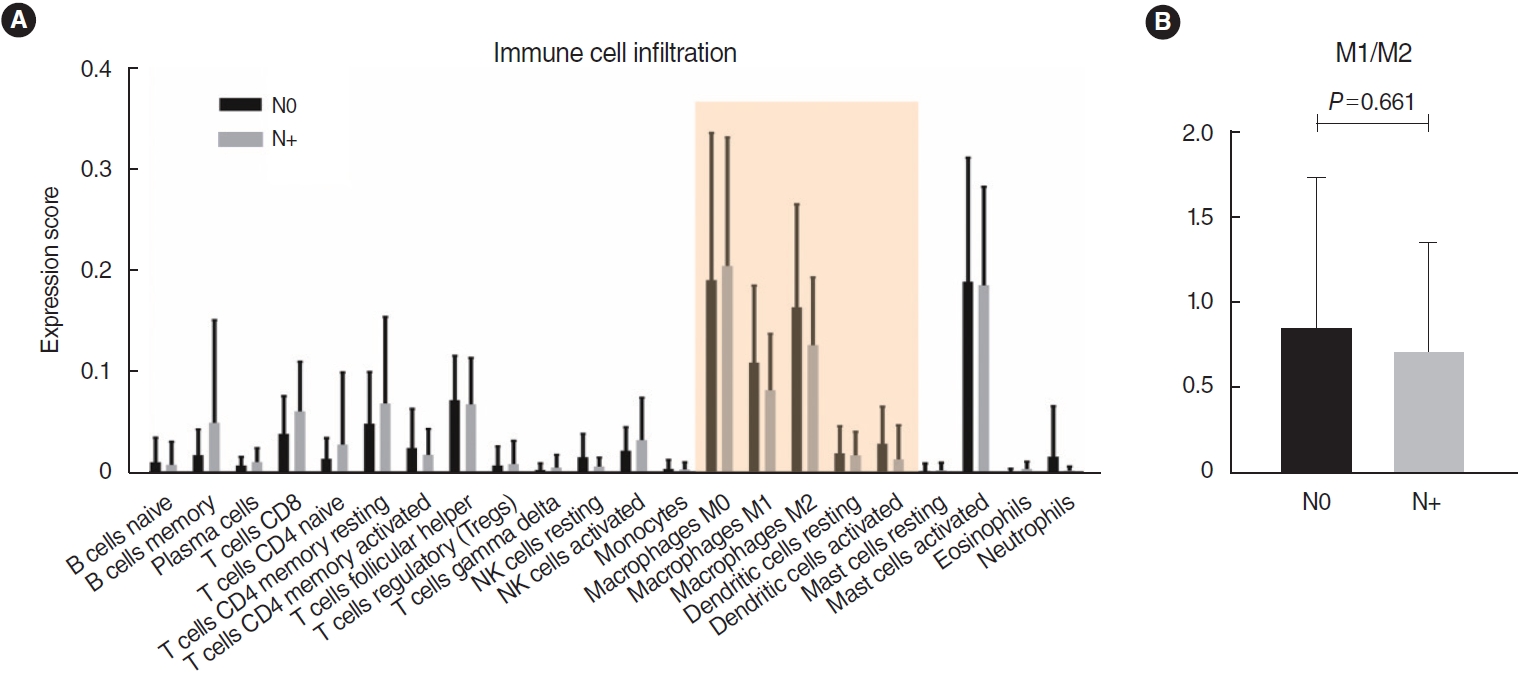
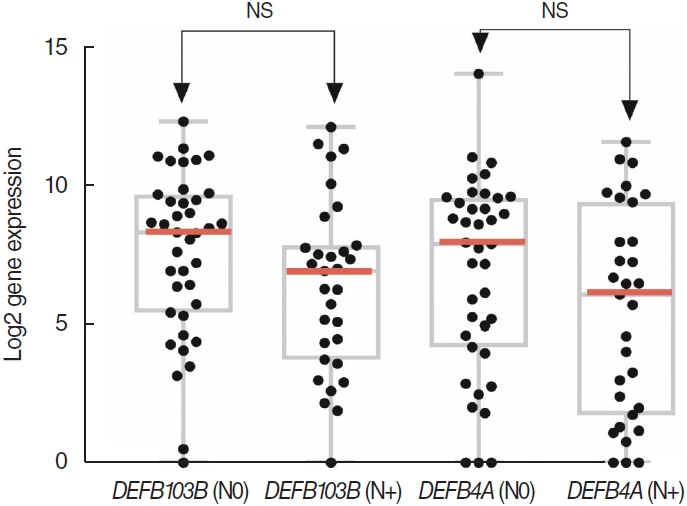
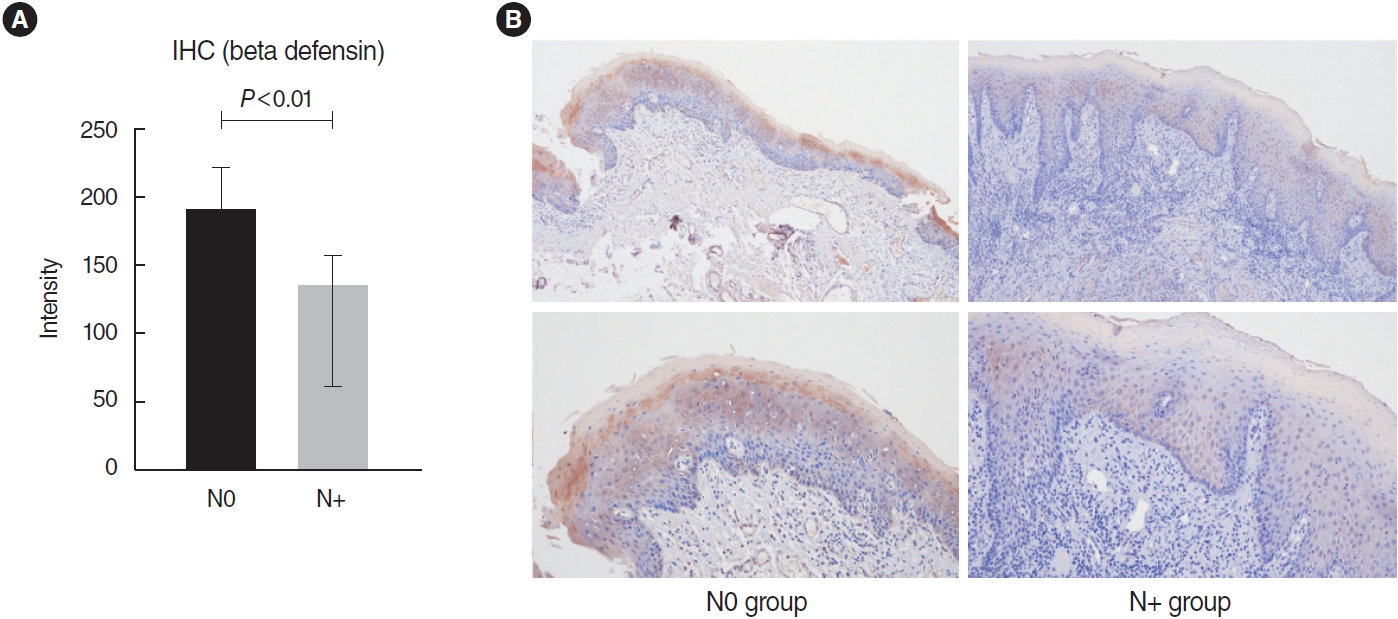
. No significant differences were found in the infiltration of 22 types of immune cells. Among a total of 51 identified DEGs, 14 genes were significantly upregulated, while 37 genes were significantly downregulated (P<0.01; fold change >2). Pathway analysis revealed significant associations with the arachidonic acid metabolism-related pathway, calcium signaling, and the muscle contraction pathway. The following DEGs were the most significantly different between the two groups: DEFB4A, SPRR2B, DEFB103B, SPRR2G, DEFB4B, and FAM25A. TCGA data showed that DEFB4A and DEFB103B were more highly expressed in the N0 group than in the N+ group, although the difference did not achieve statistical significance. Immunochemical staining of cancer tissue revealed significantly higher expression of defensin in the N0 group.
Conclusions
. Defensin (DEFB4A, DEFB103B, DEFB4B) may be a novel biomarker for early regional metastasis in T1/2 tongue cancer.
Figure
Cited by 2 articles
-
Prediction of Occult Lymph Node Metastasis in Early Tongue Cancer
Minsu Kwon
Clin Exp Otorhinolaryngol. 2022;15(4):297-298. doi: 10.21053/ceo.2022.01445.Gene Expression Alteration by Non-thermal Plasma-Activated Media Treatment in Radioresistant Head and Neck Squamous Cell Carcinoma
Sicong Zheng, Yudan Piao, Seung-Nam Jung, Chan Oh, Mi Ae Lim, QuocKhanh Nguyen, Shan Shen, Se-Hee Park, Shengzhe Cui, Shuyu Piao, Young Il Kim, Ji Won Kim, Ho-Ryun Won, Jae Won Chang, Yujuan Shan, Lihua Liu, Bon Seok Koo
Clin Exp Otorhinolaryngol. 2025;18(1):73-87. doi: 10.21053/ceo.2024.00238.
Reference
-
1. Okura M, Aikawa T, Sawai NY, Iida S, Kogo M. Decision analysis and treatment threshold in a management for the N0 neck of the oral cavity carcinoma. Oral Oncol. 2009; Oct. 45(10):908–11.
Article2. Fasunla AJ, Greene BH, Timmesfeld N, Wiegand S, Werner JA, Sesterhenn AM. A meta-analysis of the randomized controlled trials on elective neck dissection versus therapeutic neck dissection in oral cavity cancers with clinically node-negative neck. Oral Oncol. 2011; May. 47(5):320–4.
Article3. Kurokawa H, Yamashita Y, Takeda S, Zhang M, Fukuyama H, Takahashi T. Risk factors for late cervical lymph node metastases in patients with stage I or II carcinoma of the tongue. Head Neck. 2002; Jul. 24(8):731–6.
Article4. Keski-Santti H, Atula T, Tornwall J, Koivunen P, Makitie A. Elective neck treatment versus observation in patients with T1/T2 N0 squamous cell carcinoma of oral tongue. Oral Oncol. 2006; Jan. 42(1):96–101.5. Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015; Mar. 12(4):357–60.
Article6. Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015; Feb. 33(3):290–5.
Article7. Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, stringtie and ballgown. Nat Protoc. 2016; Aug. 11(9):1650–67.
Article8. Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015; Mar. 12(5):453–7.
Article9. van’t Veer LJ, Bernards R. Enabling personalized cancer medicine through analysis of gene-expression patterns. Nature. 2008; Apr. 452(7187):564–70.
Article10. Chung CH, Parker JS, Karaca G, Wu J, Funkhouser WK, Moore D, et al. Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell. 2004; May. 5(5):489–500.
Article11. Watanabe H, Mogushi K, Miura M, Yoshimura R, Kurabayashi T, Shibuya H, et al. Prediction of lymphatic metastasis based on gene expression profile analysis after brachytherapy for early-stage oral tongue carcinoma. Radiother Oncol. 2008; May. 87(2):237–42.
Article12. van Hooff SR, Leusink FK, Roepman P, Baatenburg de Jong RJ, Speel EJ, van den Brekel MW, et al. Validation of a gene expression signature for assessment of lymph node metastasis in oral squamous cell carcinoma. J Clin Oncol. 2012; Nov. 30(33):4104–10.
Article13. Raj PA, Dentino AR. Current status of defensins and their role in innate and adaptive immunity. FEMS Microbiol Lett. 2002; Jan. 206(1):9–18.
Article14. Oppenheim JJ, Biragyn A, Kwak LW, Yang D. Roles of antimicrobial peptides such as defensins in innate and adaptive immunity. Ann Rheum Dis. 2032; Nov. 62 Suppl 2(Suppl 2):ii17–21.
Article15. Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol. 2003; Sep. 3(9):710–20.
Article16. Yang D, Biragyn A, Hoover DM, Lubkowski J, Oppenheim JJ. Multiple roles of antimicrobial defensins, cathelicidins, and eosinophil-derived neurotoxin in host defense. Annu Rev Immunol. 2004; Jan. 22:181–215.
Article17. Schutte BC, Mitros JP, Bartlett JA, Walters JD, Jia HP, Welsh MJ, et al. Discovery of five conserved beta-defensin gene clusters using a computational search strategy. Proc Natl Acad Sci U S A. 2002; Feb. 99(4):2129–33.18. Wu Z, Hoover DM, Yang D, Boulegue C, Santamaria F, Oppenheim JJ, et al. Engineering disulfide bridges to dissect antimicrobial and chemotactic activities of human beta-defensin 3. Proc Natl Acad Sci U S A. 2003; Jul. 100(15):8880–5.19. Sass V, Schneider T, Wilmes M, Korner C, Tossi A, Novikova N, et al. Human beta-defensin 3 inhibits cell wall biosynthesis in Staphylococci. Infect Immun. 2010; Jun. 78(6):2793–800.
Article20. Donald CD, Sun CQ, Lim SD, Macoska J, Cohen C, Amin MB, et al. Cancer-specific loss of beta-defensin 1 in renal and prostatic carcinomas. Lab Invest. 2003; Apr. 83(4):501–5.
Article21. Gambichler T, Skrygan M, Huyn J, Bechara FG, Sand M, Altmeyer P, et al. Pattern of mRNA expression of beta-defensins in basal cell carcinoma. BMC Cancer. 2006; Jun. 6:163.22. Bullard RS, Gibson W, Bose SK, Belgrave JK, Eaddy AC, Wright CJ, et al. Functional analysis of the host defense peptide human beta defensin-1: new insight into its potential role in cancer. Mol Immunol. 2008; Feb. 45(3):839–48.
Article23. Bissell J, Joly S, Johnson GK, Organ CC, Dawson D, McCray PB Jr, et al. Expression of beta-defensins in gingival health and in periodontal disease. J Oral Pathol Med. 2004; May. 33(5):278–85.
Article24. Wenghoefer M, Pantelis A, Dommisch H, Reich R, Martini M, Allam JP, et al. Decreased gene expression of human beta-defensin-1 in the development of squamous cell carcinoma of the oral cavity. Int J Oral Maxillofac Surg. 2008; Jul. 37(7):660–3.25. Han Q, Wang R, Sun C, Jin X, Liu D, Zhao X, et al. Human beta-defensin-1 suppresses tumor migration and invasion and is an independent predictor for survival of oral squamous cell carcinoma patients. PLoS One. 2014; Mar. 9(3):e91867.
Article26. Cao Y, Green K, Quattlebaum S, Milam B, Lu L, Gao D, et al. Methylated genomic loci encoding microRNA as a biomarker panel in tissue and saliva for head and neck squamous cell carcinoma. Clin Epigenetics. 2018; Apr. 10:43.
Article27. Gualtero DF, Suarez Castillo A. Biomarkers in saliva for the detection of oral squamous cell carcinoma and their potential use for early diagnosis: a systematic review. Acta Odontol Scand. 2016; 74(3):170–7.
Article28. Svensson D, Aidoukovitch A, Anders E, Agerberth B, Andersson F, Ekblad E, et al. The host defense peptide LL-37 is detected in human parotid and submandibular/sublingual saliva and expressed in glandular neutrophils. Eur J Oral Sci. 2018; Apr. 126(2):93–100.
Article29. Shuyi Y, Feng W, Jing T, Hongzhang H, Haiyan W, Pingping M, et al. Human beta-defensin-3 (hBD-3) upregulated by LPS via epidermal growth factor receptor (EGFR) signaling pathways to enhance lymphatic invasion of oral squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2011; Nov. 112(5):616–25.
Article30. Shi N, Jin F, Zhang X, Clinton SK, Pan Z, Chen T. Overexpression of human β-defensin 2 promotes growth and invasion during esophageal carcinogenesis. Oncotarget. 2014; Nov. 5(22):11333–44.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Correlation Between Expression of p53, Bcl-2 Protein and Ki-67 Labelling Index and Lymph Node Metastasis in Early Gastric Cancer
- Sentinel Lymph Node Centered Selective Neck Dissection Does Not Have Benefits Over Supraomohyoid Neck Dissection in Patients with cT1T2N0 Tongue Cancer
- Expression of Oncogenes in Gastric Cancer: As a Correlation between Oncogene Expression and Prognosis
- Analysis of Influencing Factors on Cervical Lymph Node Metastasis in Early Oral Tongue Cancer
- Expression and clinical significance of defensin alpha 6 in colorectal cancer