Healthc Inform Res.
2022 Oct;28(4):364-375. 10.4258/hir.2022.28.4.364.
Machine Learning Model for the Prediction of Hemorrhage in Intensive Care Units
- Affiliations
-
- 1Department of Biomedical Informatics, Ajou University School of Medicine, Suwon, Korea
- 2Division of Pulmonology, Department of Internal Medicine, Wonkwang University Hospital, Iksan, Korea
- 3Department of Biomedical Engineering, College of Electronics and Information, Kyung Hee University, Yongin, Korea
- 4Department of Biomedical Systems Informatics, Yonsei University College of Medicine, Yongin, Korea
- 5Center for Digital Health, Yongin Severance Hospital, Yonsei University Health System, Yongin, Korea
- 6BUD.on Inc., Jeonju, Korea
- KMID: 2535811
- DOI: http://doi.org/10.4258/hir.2022.28.4.364
Abstract
Objectives
Early hemorrhage detection in intensive care units (ICUs) enables timely intervention and reduces the risk of irreversible outcomes. In this study, we aimed to develop a machine learning model to predict hemorrhage by learning the patterns of continuously changing, real-world clinical data.
Methods
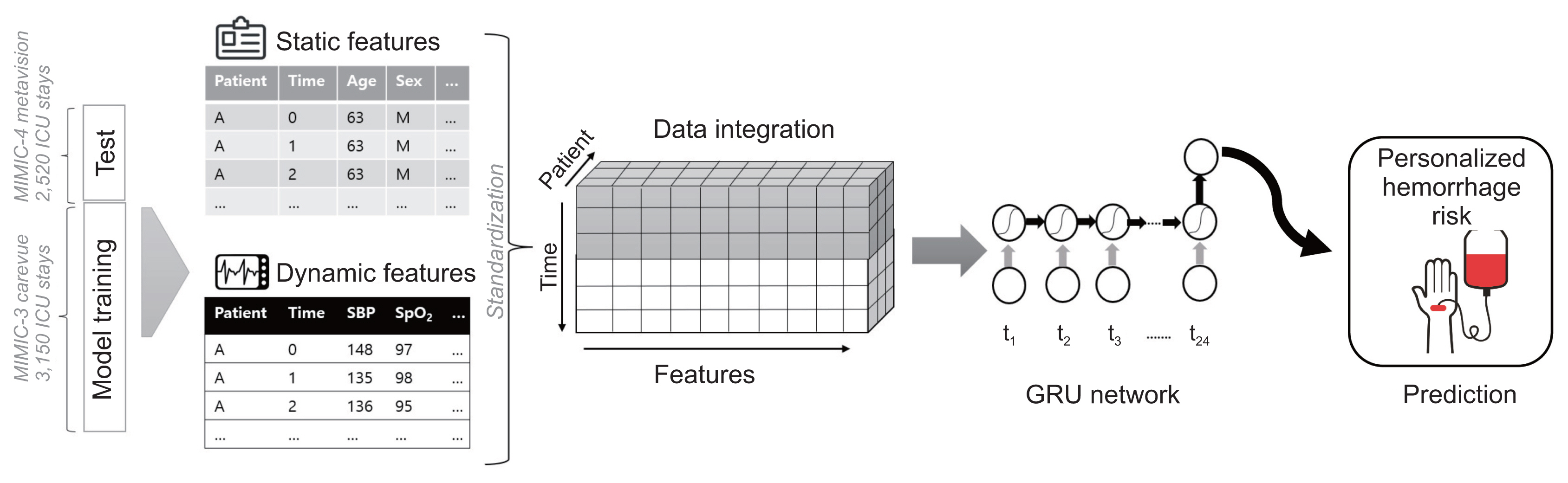
We used the Medical Information Mart for Intensive Care databases (MIMIC-III and MIMIC-IV). A recurrent neural network was used to predict severe hemorrhage in the ICU. We developed three machine learning models with an increasing number of input features and levels of complexity: model 1 (11 features), model 2 (18 features), and model 3 (27 features). MIMIC-III was used for model training, and MIMIC-IV was split for internal validation. Using the model with the highest performance, external verification was performed using data from a subgroup extracted from the eICU Collaborative Research Database.
Results
We included 5,670 ICU admissions, with 3,150 in the training set and 2,520 in the internal test set. A positive correlation was found between model complexity and performance. As a measure of performance, three models developed with an increasing number of features showed area under the receiver operating characteristic (AUROC) curve values of 0.61–0.94 according to the range of input data. In the subgroup extracted from the eICU database for external validation, an AUROC value of 0.74 was observed.
Conclusions
Machine learning models that rely on real clinical data can be used to predict patients at high risk of bleeding in the ICU.
Figure
Reference
-
References
1. Despotis G, Avidan M, Eby C. Prediction and management of bleeding in cardiac surgery. J Thromb Haemost. 2009; 7(Suppl 1):111–7. https://doi.org/10.1111/j.1538-7836.2009.03412.x .
Article2. Ferraris VA, Hochstetler M, Martin JT, Mahan A, Saha SP. Blood transfusion and adverse surgical outcomes: the good and the bad. Surgery. 2015; 158(3):608–17. https://doi.org/10.1016/j.surg.2015.02.027 .
Article3. Rao SV, Jollis JG, Harrington RA, Granger CB, Newby LK, Armstrong PW, et al. Relationship of blood transfusion and clinical outcomes in patients with acute coronary syndromes. JAMA. 2004; 292(13):1555–62. https://doi.org/10.1001/jama.292.13.1555 .
Article4. Cook DJ, Griffith LE, Walter SD, Guyatt GH, Meade MO, Heyland DK, et al. The attributable mortality and length of intensive care unit stay of clinically important gastrointestinal bleeding in critically ill patients. Crit Care. 2001; 5(6):368–75. https://doi.org/10.1186/cc1071 .
Article5. Pirie L, McClelland DB, Franklin IM. EU optimal blood use project partners and project management team. The EU optimal blood use project. Transfus Clin Biol. 2007; 14(6):499–503. https://doi.org/10.1016/j.tracli.2008.03.005 .
Article6. Mitterecker A, Hofmann A, Trentino KM, Lloyd A, Leahy MF, Schwarzbauer K, et al. Machine learning-based prediction of transfusion. Transfusion. 2020; 60(9):1977–86. https://doi.org/10.1111/trf.15935 .
Article7. Janett RS, Yeracaris PP. Electronic medical records in the American Health System: challenges and lessons learned. Cien Saude Colet. 2020; 25(4):1293–304. https://doi.org/10.1590/1413-81232020254.28922019 .
Article8. Carra G, Salluh JIF, da Silva Ramos FJ, Meyfroidt G. Data-driven ICU management: using big data and algorithms to improve outcomes. J Crit Care. 2020; 60:300–4. https://doi.org/10.1016/j.jcrc.2020.09.002 .
Article9. Celi LA, Mark RG, Stone DJ, Montgomery RA. “Big data” in the intensive care unit: closing the data loop. Am J Respir Crit Care Med. 2013; 187(11):1157–60. https://doi.org/10.1164/rccm.201212-2311ed .
Article10. Chen JH, Asch SM. Machine learning and prediction in medicine: beyond the peak of inflated expectations. N Engl J Med. 2017; 376(26):2507–9. https://doi.org/10.1056/nejmp1702071 .
Article11. Gombotz H, Knotzer H. Preoperative identification of patients with increased risk for perioperative bleeding. Curr Opin Anaesthesiol. 2013; 26(1):82–90. https://doi.org/10.1097/aco.0b013e32835b9a23 .
Article12. Thorson CM, Ryan ML, Van Haren RM, Pereira R, Olloqui J, Otero CA, et al. Change in hematocrit during trauma assessment predicts bleeding even with ongoing fluid resuscitation. Am Surg. 2013; 79(4):398–406. https://doi.org/10.1177/000313481307900430 .
Article13. Bruns B, Lindsey M, Rowe K, Brown S, Minei JP, Gentilello LM, et al. Hemoglobin drops within minutes of injuries and predicts need for an intervention to stop hemorrhage. J Trauma. 2007; 63(2):312–5. https://doi.org/10.1097/ta.0b013e31812389d6 .
Article14. Thorson CM, Van Haren RM, Ryan ML, Pereira R, Olloqui J, Guarch GA, et al. Admission hematocrit and transfusion requirements after trauma. J Am Coll Surg. 2013; 216(1):65–73. https://doi.org/10.1016/j.jamcollsurg.2012.09.011 .
Article15. Koshkareva YA, Cohen M, Gaughan JP, Callanan V, Szeremeta W. Utility of preoperative hematologic screening for pediatric adenotonsillectomy. Ear Nose Throat J. 2012; 91(8):346–56. https://doi.org/10.1177/014556131209100809 .
Article16. Shumborski S, Gooden B, Salmon LJ, O’Sullivan M, Pinczewski LA, Roe JP, et al. Utility of preoperative blood screening before hip and knee arthroplasty. ANZ J Surg. 2020; 90(3):350–4. https://doi.org/10.1111/ans.15676 .
Article17. Haberle J. Clinical and biochemical aspects of primary and secondary hyperammonemic disorders. Arch Biochem Biophys. 2013; 536(2):101–8. https://doi.org/10.1016/j.abb.2013.04.009 .
Article18. Al-Naamani K, Alzadjali N, Barkun AN, Fallone CA. Does blood urea nitrogen level predict severity and high-risk endoscopic lesions in patients with nonvariceal upper gastrointestinal bleeding? Can J Gastroenterol. 2008; 22(4):399–403. https://doi.org/10.1155/2008/207850 .
Article19. Tomizawa M, Shinozaki F, Hasegawa R, Shirai Y, Motoyoshi Y, Sugiyama T, et al. Patient characteristics with high or low blood urea nitrogen in upper gastrointestinal bleeding. World J Gastroenterol. 2015; 21(24):7500–5. https://doi.org/10.3748/wjg.v21.i24.7500 .
Article20. Meyer A, Zverinski D, Pfahringer B, Kempfert J, Kuehne T, Sundermann SH, et al. Machine learning for real-time prediction of complications in critical care: a retrospective study. Lancet Respir Med. 2018; 6(12):905–14. https://doi.org/10.1016/s2213-2600(18)30300-x .
Article21. Bonde A, Varadarajan KM, Bonde N, Troelsen A, Muratoglu OK, Malchau H, et al. Assessing the utility of deep neural networks in predicting postoperative surgical complications: a retrospective study. Lancet Digit Health. 2021; 3(8):e471–85. https://doi.org/10.1016/s2589-7500(21)00084-4 .
Article22. Levi R, Carli F, Arevalo AR, Altinel Y, Stein DJ, Naldini MM, et al. Artificial intelligence-based prediction of transfusion in the intensive care unit in patients with gastrointestinal bleeding. BMJ Health Care Inform. 2021; 28(1):e100245. https://doi.org/10.1136/bmjhci-2020-100245 .
Article23. Johnson AE, Pollard TJ, Shen L, Lehman LW, Feng M, Ghassemi M, et al. MIMIC-III, a freely accessible critical care database. Sci Data. 2016; 3:160035. https://doi.org/10.1038/sdata.2016.35 .
Article24. Graves A. Supervised sequence labelling with recurrent neural networks. Heidelberg, Germany: Springer;2012. https://doi.org/10.1007/978-3-642-24797-2 .
Article25. Cho K, van Merrienboer B, Gulcehre C, Bahdanau D, Bougares F, Schwenk H, et al. Learning phrase representations using RNN encoder-decoder for statistical machine translation [Internet]. Ithaca (NY): arXiv.org;2014. [cited at 2022 Sep 30]. Available from: https://arxiv.org/abs/1406.1078 .26. Pollard TJ, Johnson AE, Raffa JD, Celi LA, Mark RG, Badawi O. The eICU Collaborative Research Database, a freely available multi-center database for critical care research. Sci Data. 2018; 5:180178. https://doi.org/10.1038/sdata.2018.178 .
Article27. Zhu W, He W, Guo L, Wang X, Hong K. The HASBLED Score for predicting major bleeding risk in anti-coagulated patients with atrial fibrillation: a systematic review and meta-analysis. Clin Cardiol. 2015; 38(9):555–61. https://doi.org/10.1002/clc.22435 .
Article28. Bento D, Marques N, Azevedo P, Guedes J, Bispo J, Silva D, et al. CRUSADE: is it still a good score to predict bleeding in acute coronary syndrome? Rev Port Cardiol (Engl Ed). 2018; 37(11):889–97. https://doi.org/10.1016/j.repc.2018.02.008 .
Article29. Yildirim E, Uku O, Bilen MN, Secen O. Performance of HAS-BLED and CRUSADE risk scores for the prediction of haemorrhagic events in patients with stable coronary artery disease. Cardiovasc J Afr. 2019; 30(4):198–202. https://doi.org/10.5830/cvja-2019-014 .
Article30. De Pasquale M, Moss TJ, Cerutti S, Calland JF, Lake DE, Moorman JR, et al. Hemorrhage prediction models in surgical intensive care: bedside monitoring data adds information to lab values. IEEE J Biomed Health Inform. 2017; 21(6):1703–10. https://doi.org/10.1109/jbhi.2017.2653849 .
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Machine Learning vs. Statistical Model for Prediction Modelling: Application in Medical Imaging Research
- A Comparison of Intensive Care Unit Mortality Prediction Models through the Use of Data Mining Techniques
- Hierarchical Genetic Algorithm and Fuzzy Radial Basis Function Networks for Factors Influencing Hospital Length of Stay Outliers
- An improved machine learning model for calculation of intraocular lens power during cataract surgery in Republic of Korea: development
- Artificial intelligence, machine learning, and deep learning in women’s health nursing