J Pathol Transl Med.
2022 Mar;56(2):92-96. 10.4132/jptm.2021.10.15.
An unusual case of microsatellite instability–high/deficient mismatch repair (MSI-H/dMMR) diffuse large B-cell lymphoma revealed by targeted gene sequencing
- Affiliations
-
- 1Department of Pathology, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea
- 2Center for Precision Medicine, Seoul National University Hospital, Seoul, Korea
- 3Cancer Research Institute, Seoul National University, Seoul, Korea
- KMID: 2527193
- DOI: http://doi.org/10.4132/jptm.2021.10.15
Abstract
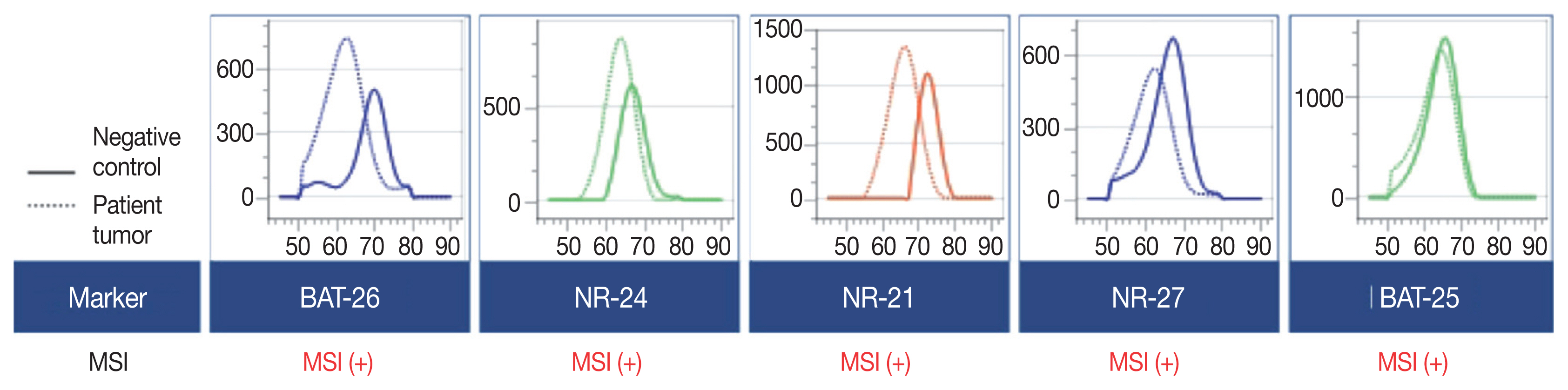
- Microsatellite instability-high/deficient mismatch repair (MSI-H/dMMR) status has been approved as a tissue-agnostic biomarker for immune checkpoint inhibitor therapy in patients with solid tumors. We report the case of an MSI-H/dMMR diffuse large B-cell lymphoma (DLBCL) identified by targeted gene sequencing (TGS). A 90-year-old female who presented with vaginal bleeding and a large mass in the upper vagina was diagnosed with germinal center-B-cell-like DLBCL, which recurred at the uterine cervix at 9 months after chemotherapy. Based on TGS of 121 lymphoma-related genes and the LymphGen algorithm, the tumor was classified genetically as DLBCL of EZB subtype. Mutations in multiple genes, including frequent frameshift mutations, were detected by TGS and further suggested MSI. The MSI-H/dMMR and loss of MLH1 and PMS2 expression were determined in MSI-fragment analysis, MSI real-time polymerase chain reaction, and immunohistochemical tests. This case demonstrates the potential diagnostic and therapeutic utility of lymphoma panel sequencing for DLBCL with MSI-H/dMMR.
Keyword
Figure
Reference
-
References
1. Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004; 96:261–8.2. Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017; 357:409–13.3. Marcus L, Lemery SJ, Keegan P, Pazdur R. FDA approval summary: pembrolizumab for the treatment of microsatellite instability-high solid tumors. Clin Cancer Res. 2019; 25:3753–8.
Article4. Bonneville R, Krook MA, Kautto EA, et al. Landscape of microsatellite instability across 39 cancer types. JCO Precis Oncol. 2017; 2017:PO.17.00073.
Article5. Middha S, Zhang L, Nafa K, et al. Reliable pan-cancer microsatellite instability assessment by using targeted next-generation sequencing data. JCO Precis Oncol. 2017; 2017:PO.17.00084.
Article6. Albayrak A, Garrido-Castro AC, Giannakis M, et al. Clinical pan-cancer assessment of mismatch repair deficiency using tumor-only, targeted next-generation sequencing. JCO Precis Oncol. 2020; 4:1084–97.
Article7. Trabucco SE, Gowen K, Maund SL, et al. A novel next-generation sequencing approach to detecting microsatellite instability and pan-tumor characterization of 1000 microsatellite instability-high cases in 67,000 patient samples. J Mol Diagn. 2019; 21:1053–66.
Article8. Duval A, Raphael M, Brennetot C, et al. The mutator pathway is a feature of immunodeficiency-related lymphomas. Proc Natl Acad Sci U S A. 2004; 101:5002–7.
Article9. Tian T, Li J, Xue T, Yu B, Li X, Zhou X. Microsatellite instability and its associations with the clinicopathologic characteristics of diffuse large B-cell lymphoma. Cancer Med. 2020; 9:2330–42.
Article10. Wright GW, Huang DW, Phelan JD, et al. A probabilistic classification tool for genetic subtypes of diffuse large B cell lymphoma with therapeutic implications. Cancer Cell. 2020; 37:551–68.
Article11. Suraweera N, Duval A, Reperant M, et al. Evaluation of tumor microsatellite instability using five quasimonomorphic mononucleotide repeats and pentaplex PCR. Gastroenterology. 2002; 123:1804–11.
Article12. Hause RJ, Pritchard CC, Shendure J, Salipante SJ. Classification and characterization of microsatellite instability across 18 cancer types. Nat Med. 2016; 22:1342–50.
Article13. Armand P, Rodig S, Melnichenko V, et al. Pembrolizumab in relapsed or refractory primary mediastinal large B-cell lymphoma. J Clin Oncol. 2019; 37:3291–9.
Article14. Kuruvilla J, Ramchandren R, Santoro A, et al. Pembrolizumab versus brentuximab vedotin in relapsed or refractory classical Hodgkin lymphoma (KEYNOTE-204): an interim analysis of a multicentre, randomised, open-label, phase 3 study. Lancet Oncol. 2021; 22:512–24.
Article15. Smith SD, Till BG, Shadman MS, et al. Pembrolizumab with R-CHOP in previously untreated diffuse large B-cell lymphoma: potential for biomarker driven therapy. Br J Haematol. 2020; 189:1119–26.
Article16. de Miranda NF, Peng R, Georgiou K, et al. DNA repair genes are selectively mutated in diffuse large B cell lymphomas. J Exp Med. 2013; 210:1729–42.
Article17. Dubois S, Viailly PJ, Mareschal S, et al. Next-generation sequencing in diffuse large B-cell lymphoma highlights molecular divergence and therapeutic opportunities: a LYSA study. Clin Cancer Res. 2016; 22:2919–28.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Comprehensive Study of Microsatellite Instability Testing and Its Comparison With Immunohistochemistry in Gastric Cancers
- Microsatellite Instability and Mismatch Repair Protein (hMLH1, hMSH2) Expression in Intrahepatic Cholangiocarcinoma
- Recent Progress in Immunotherapy for Metastatic Colorectal Cancer
- Immune check point inhibitors in BRAF mutated metastatic colorectal cancer: A review
- Microsatellite Instability in Gastric B-cell Lymphoma