Lab Med Online.
2021 Jan;11(1):17-24. 10.47429/lmo.2021.11.1.17.
NF1 Variant Spectrum in Korean Patients with Neurofibromatosis Type 1 Disorder
- Affiliations
-
- 1Department of Laboratory Medicine, College of Medicine, The Catholic University of Korea, Seoul, Korea
- 2Catholic Genetic Laboratory Center, Seoul St. Mary’s Hospital, College of Medicine, The Catholic University of Korea, Seoul, Korea
- KMID: 2525776
- DOI: http://doi.org/10.47429/lmo.2021.11.1.17
Abstract
- Background
Neurofibromatosis type 1 (NF1) is one of the most common autosomal dominant disorders in humans, and many different variants in the NF1 gene have been observed. The aim of this study was to investigate the genetic variant spectrum of NF1patients in Korea.
Methods
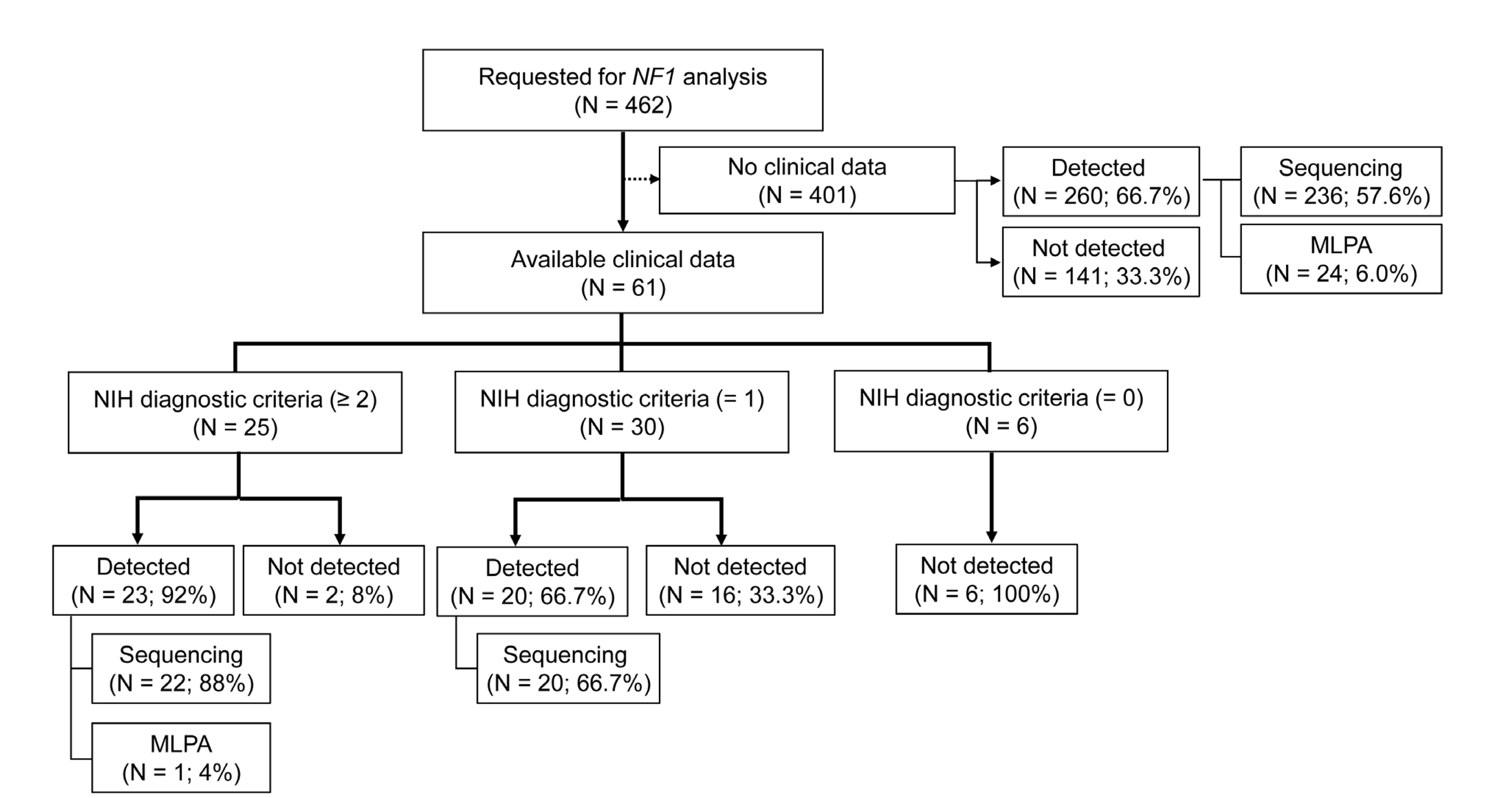
A total of 462 cases were enrolled for NF1 analysis at Seoul St. Mary’s Hospital. NF1 was analyzed through Sanger sequencing of messenger RNA (mRNA) and genomic DNA (gDNA), and/or multiplex ligation-dependent probe amplification (MLPA) analysis.
Results
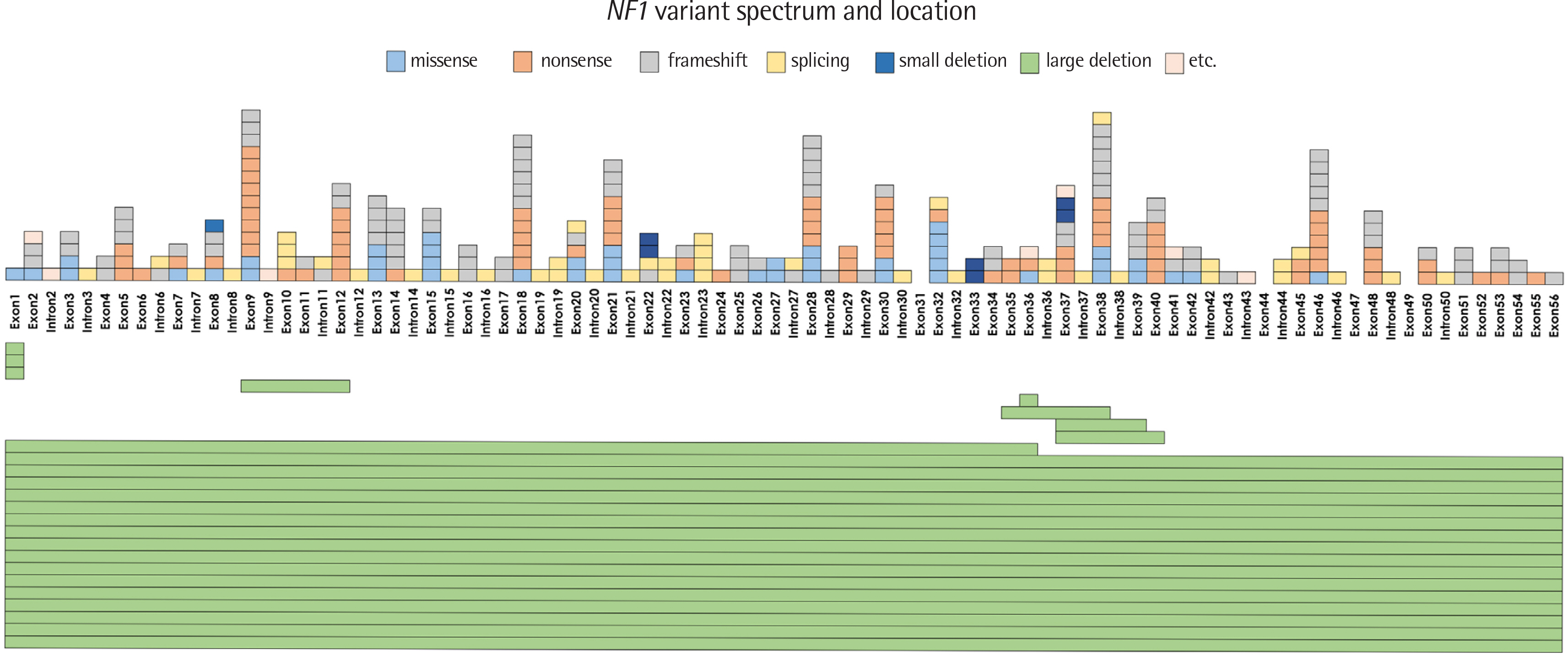
We identified 231 types of NF1 variants in 303 out of the 462 patients (65.6%). Of these, 217 variants were classified as pathogenic or likely pathogenic, and 48.4% (N = 106) of these were novel changes. Truncating variants encompassing frameshift and nonsense variants were most commonly observed (135 types of variants in 177 patients), followed by splicing defect variants (39 types in 42 patients) and missense variants (36 types in 44 patients). There were 8 distinctive large deletions in 25 patients, detected via MLPA analysis. Interestingly, 4 cases showed aberrant transcripts that were identified through mRNA Sanger sequencing. The frequency of NF1 mutation detected was significantly higher according to the number of satisfying National Institutes of Health (NIH) diagnostic criteria.
Conclusions
This study revealed a wide spectrum of NF1 variants in Korean NF1 patients. A comprehensive analytical strategy that combines mRNA and gDNA analyses and MLPA is required to detect sequence variants. Additionally, it is important to define the effect of newly detected variants on the clinical course.
Keyword
Figure
Reference
-
1. Reynolds RM, Browning GG, Nawroz I, Campbell IW. 2003; Von Recklinghausen's neurofibromatosis: neurofibromatosis type 1. Lancet. 361:1552–4. DOI: 10.1016/S0140-6736(03)13166-2.2. Jett K, Friedman JM. 2010; Clinical and genetic aspects of neurofibromatosis 1. Genet Med. 12:1–11. DOI: 10.1097/GIM.0b013e3181bf15e3. PMID: 20027112.3. Valero MC, Martín Y, Hernández-Imaz E, Marina Hernández A, Meleán G, Valero AM, et al. 2011; A highly sensitive genetic protocol to detect NF1 mutations. J Mol Diagn. 13:113–22. DOI: 10.1016/j.jmoldx.2010.09.002. PMID: 21354044. PMCID: PMC3128626.4. Messiaen L, Wimmer K, editors. 2008. NF1 mutational spectrum. Karger;Basel: p. 63–77. DOI: 10.1159/000126545.5. Messiaen L, Wimmer K. 2012. Mutation analysis of the NF1 gene by cDNA-based sequencing of the coding region. Advances in neurofibromatosis research. Nova Science Publishers, Inc..6. Evans DG, Bowers N, Burkitt-Wright E, Miles E, Garg S, Scott-Kitching V, et al. 2016; Comprehensive RNA analysis of the NF1 gene in classically affected NF1 affected individuals meeting NIH criteria has high sensitivity and mutation negative testing is reassuring in isolated cases with pigmentary features only. EBioMedicine. 7:212–20. DOI: 10.1016/j.ebiom.2016.04.005. PMID: 27322474. PMCID: PMC4909377.7. Stenson PD, Mort M, Ball EV, Evans K, Hayden M, Heywood S, et al. 2017; The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum Genet. 136:665–77. DOI: 10.1007/s00439-017-1779-6. PMID: 28349240. PMCID: PMC5429360.8. Messiaen LM, Callens T, Mortier G, Beysen D, Vandenbroucke I, Van Roy N, et al. 2000; Exhaustive mutation analysis of the NF1 gene allows identification of 95% of mutations and reveals a high frequency of unusual splicing defects. Hum Mutat. 15:541–55. DOI: 10.1002/1098-1004(200006)15:6<541::AID-HUMU6>3.0.CO;2-N.9. Jo YH, Kim HO, Song HR, Yoon KS. 2014; Identification of the NF1 gene mutation in Korean families with neurofibromatosis type 1. Genes & Genomics. 36:11–5. DOI: 10.1007/s13258-013-0132-2.10. Jang MA, Kim YE, Kim SK, Lee MK, Kim JW, Ki CS. 2016; Identification and characterization of NF1 splicing mutations in Korean patients with neurofibromatosis type 1. J Hum Genet. 61:705–9. DOI: 10.1038/jhg.2016.33. PMID: 27074763.11. Jeong SY, Park SJ, Kim HJ. 2006; The spectrum of NF1 mutations in Korean patients with neurofibromatosis type 1. J Korean Med Sci. 21:107–12. DOI: 10.3346/jkms.2006.21.1.107. PMID: 16479075. PMCID: PMC2733956.12. Kim MJ, Cheon CK. 2014; Neurofibromatosis type 1: a single center's experience in Korea. Korean J Pediatr. 57:410–5. DOI: 10.3345/kjp.2014.57.9.410. PMID: 25324867. PMCID: PMC4198956.13. Ko JM, Sohn YB, Jeong SY, Kim HJ, Messiaen LM. 2013; Mutation spectrum of NF1 and clinical characteristics in 78 Korean patients with neurofibromatosis type 1. Pediatr Neurol. 48:447–53. DOI: 10.1016/j.pediatrneurol.2013.02.004. PMID: 23668869.14. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. 2015; Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 17:405–24. DOI: 10.1038/gim.2015.30. PMID: 25741868. PMCID: PMC4544753.15. De Luca A, Bottillo I, Dasdia MC, Morella A, Lanari V, Bernardini L, et al. 2007; Deletions of NF1 gene and exons detected by multiplex ligation-dependent probe amplification. J Med Genet. 44:800–8. DOI: 10.1136/jmg.2007.053785. PMID: 18055911. PMCID: PMC2652822.16. Ars E, Kruyer H, Morell M, Pros E, Serra E, Ravella A, et al. 2003; Recurrent mutations in the NF1 gene are common among neurofibromatosis type 1 patients. J Med Genet. 40:e82. DOI: 10.1136/jmg.40.6.e82. PMID: 12807981. PMCID: PMC1735494.17. Lee MJ, Su YN, You HL, Chiou SC, Lin LC, Yang CC, et al. 2006; Identification of forty-five novel and twenty-three known NF1 mutations in Chinese patients with neurofibromatosis type 1. Hum Mutat. 27:832. DOI: 10.1002/humu.9446. PMID: 16835897.18. Griffiths S, Thompson P, Frayling I, Upadhyaya M. 2007; Molecular diagnosis of neurofibromatosis type 1: 2 years experience. Fam Cancer. 6:21–34. DOI: 10.1007/s10689-006-9001-3. PMID: 16944272.19. Sabbagh A, Pasmant E, Imbard A, Luscan A, Soares M, Blanché H, et al. 2013; NF1 molecular characterization and neurofibromatosis type I genotype-phenotype correlation: the French experience. Hum Mutat. 34:1510–8. DOI: 10.1002/humu.22392. PMID: 23913538.20. Maruoka R, Takenouchi T, Torii C, Shimizu A, Misu K, Higasa K, et al. 2014; The use of next-generation sequencing in molecular diagnosis of neurofibromatosis type 1: a validation study. Genet Test Mol Biomarkers. 18:722–35. DOI: 10.1089/gtmb.2014.0109. PMID: 25325900. PMCID: PMC4216997.21. van Minkelen R, van Bever Y, Kromosoeto JN, Withagen-Hermans CJ, Nieuwlaat A, Halley DJ, et al. 2014; A clinical and genetic overview of 18 years neurofibromatosis type 1 molecular diagnostics in the Netherlands. Clin Genet. 85:318–27. DOI: 10.1111/cge.12187. PMID: 23656349.22. Zhang J, Tong H, Fu X, Zhang Y, Liu J, Cheng R, et al. 2015; Molecular characterization of NF1 and neurofibromatosis Type 1 genotype-phenotype correlations in a Chinese population. Sci Rep. 5:11291. DOI: 10.1038/srep11291. PMID: 26056819. PMCID: PMC4460887.23. Calì F, Chiavetta V, Ruggeri G, Piccione M, Selicorni A, Palazzo D, et al. 2017; Mutation spectrum of NF1 gene in Italian patients with neurofibromatosis type 1 using Ion Torrent PGM™ platform. Eur J Med Genet. 60:93–9. DOI: 10.1016/j.ejmg.2016.11.001. PMID: 27838393.24. Wu-Chou YH, Hung TC, Lin YT, Cheng HW, Lin JL, Lin CH, et al. 2018; Genetic diagnosis of neurofibromatosis type 1: targeted next-generation sequencing with Multiple Ligation-Dependent Probe Amplification analysis. J Biomed Sci. 25:72. DOI: 10.1186/s12929-018-0474-9. PMID: 30290804. PMCID: PMC6172719.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Spectrum of NF1 Mutations in Korean Patients with Neurofibromatosis Type 1
- A Rare Case of Cardiac Neurofibroma in a Patient with Neurofibromatosis Type 1: Radiologic Findings
- Attention Deficit Hyperactivity Disorder in Neurofibromatosis Type 1: Evaluation with a Continuous Performance Test
- Early-Onset Breast Cancer in a Family with Neurofibromatosis Type 1 Associated with a Germline Mutation in BRCA1
- Lifelong Management of Neurofibromatosis 1 Patients