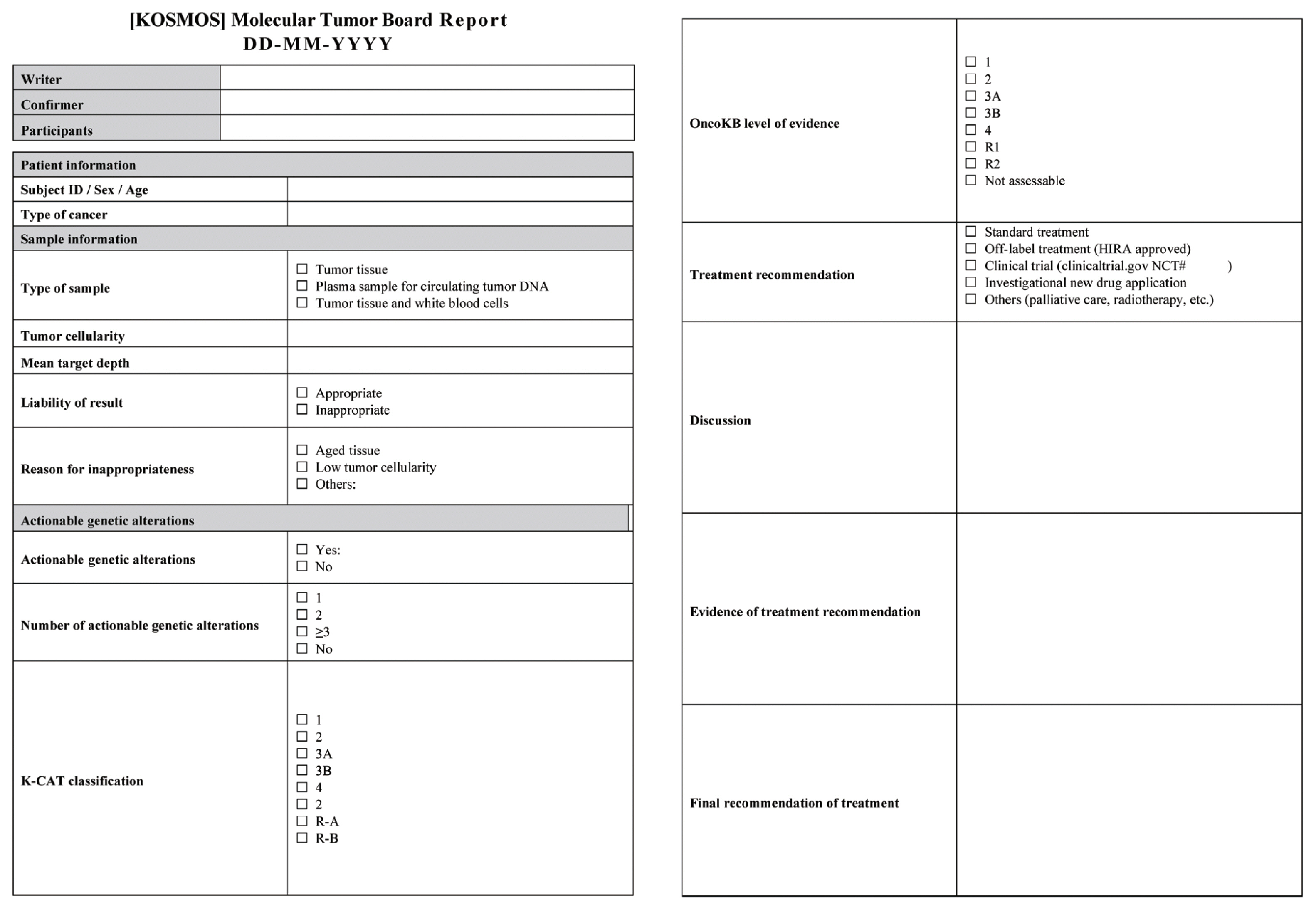
Recommendations for the Use of Next-Generation Sequencing and the Molecular Tumor Board for Patients with Advanced Cancer: A Report from KSMO and KCSG Precision Medicine Networking Group
- Affiliations
-
- 1Department of Oncology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea
- 2Department of Internal Medicine, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea
- 3Division of Medical Oncology, Yonsei Cancer Center, Yonsei University College of Medicine, Seoul, Korea
- 4Division of Hematology-Oncology, Department of Medicine, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
- 5Department of Pathology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea
- 6Department of Genomic Medicine, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea
- 7Division of Medical Oncology, Department of Internal Medicine, Gachon University Gil Medical Center, Incheon, Korea
- 8Department of Oncology/Hematology, Kyungpook National University Chilgok Hospital, Daegu, Korea
- 9Division of Medical Oncology, Department of Internal Medicine, Seoul St. Mary's Hospital, The Catholic University of Korea College of Medicine, Seoul, Korea
- 10Department of Medical Oncology, CHA Bundang Medical Center, CHA University School of Medicine, Seongnam, Korea
- 11Division of Hematology/Medical Oncology, Department of Internal Medicine, Seoul National University Bundang Hospital, Seoul National University College of Medicine, Seongnam, Korea
- 12Division of Medical Oncology, Department of Internal Medicine, Soonchunhyang University Bucheon Hospital, Bucheon, Korea
- 13Center for Colorectal Cancer, National Cancer Center, Research Institute and Hospital, Goyang, Korea
- 14Department of Internal Medicine, Hallym University Medical Center, Hallym University, Anyang, Korea
- KMID: 2524582
- DOI: http://doi.org/10.4143/crt.2021.1115
Abstract
- Next-generation sequencing (NGS) is becoming essential in the fields of precision oncology. With implementation of NGS in daily clinic, the needs for continued education, facilitated interpretation of NGS results and optimal treatment delivery based on NGS results have been addressed. Molecular tumor board (MTB) is multidisciplinary approach to keep pace with the growing knowledge of complex molecular alterations in patients with advanced solid cancer. Although guidelines for NGS use and MTB have been developed in western countries, there is limitation for reflection of Korea’s public health environment and daily clinical practice. These recommendations provide a critical guidance from NGS panel testing to final treatment decision based on MTB discussion.
Keyword
Figure
Cited by 2 articles
-
Clinical practice recommendations for the use of next-generation sequencing in patients with solid cancer: a joint report from KSMO and KSP
Miso Kim, Hyo Sup Shim, Sheehyun Kim, In Hee Lee, Jihun Kim, Shinkyo Yoon, Hyung-Don Kim, Inkeun Park, Jae Ho Jeong, Changhoon Yoo, Jaekyung Cheon, In-Ho Kim, Jieun Lee, Sook Hee Hong, Sehhoon Park, Hyun Ae Jung, Jin Won Kim, Han Jo Kim, Yongjun Cha, Sun Min Lim, Han Sang Kim, Choong-Kun Lee, Jee Hung Kim, Sang Hoon Chun, Jina Yun, So Yeon Park, Hye Seung Lee, Yong Mee Cho, Soo Jeong Nam, Kiyong Na, Sun Och Yoon, Ahwon Lee, Kee-Taek Jang, Hongseok Yun, Sungyoung Lee, Jee Hyun Kim, Wan-Seop Kim
J Pathol Transl Med. 2024;58(4):147-164. doi: 10.4132/jptm.2023.11.01.Clinical Practice Recommendations for the Use of Next-Generation Sequencing in Patients with Solid Cancer: A Joint Report from KSMO and KSP
Miso Kim, Hyo Sup Shim, Sheehyun Kim, In Hee Lee, Jihun Kim, Shinkyo Yoon, Hyung-Don Kim, Inkeun Park, Jae Ho Jeong, Changhoon Yoo, Jaekyung Cheon, In-Ho Kim, Jieun Lee, Sook Hee Hong, Sehhoon Park, Hyun Ae Jung, Jin Won Kim, Han Jo Kim, Yongjun Cha, Sun Min Lim, Han Sang Kim, Choong-kun Lee, Jee Hung Kim, Sang Hoon Chun, Jina Yun, So Yeon Park, Hye Seung Lee, Yong Mee Cho, Soo Jeong Nam, Kiyong Na, Sun Och Yoon, Ahwon Lee, Kee-Taek Jang, Hongseok Yun, Sungyoung Lee, Jee Hyun Kim, Wan-Seop Kim
Cancer Res Treat. 2024;56(3):721-742. doi: 10.4143/crt.2023.1043.
Reference
-
References
1. Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015; 372:793–5.
Article2. Biankin AV, Piantadosi S, Hollingsworth SJ. Patient-centric trials for therapeutic development in precision oncology. Nature. 2015; 526:361–70.
Article3. Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017; 23:703–13.4. Marino P, Touzani R, Perrier L, Rouleau E, Kossi DS, Zhaomin Z, et al. Cost of cancer diagnosis using next-generation sequencing targeted gene panels in routine practice: a nationwide French study. Eur J Hum Genet. 2018; 26:314–23.
Article5. Tobin NP, Foukakis T, De Petris L, Bergh J. The importance of molecular markers for diagnosis and selection of targeted treatments in patients with cancer. J Intern Med. 2015; 278:545–70.
Article6. Lee SH, Lee B, Shim JH, Lee KW, Yun JW, Kim SY, et al. Landscape of actionable genetic alterations profiled from 1,071 tumor samples in Korean cancer patients. Cancer Res Treat. 2019; 51:211–22.
Article7. Grellety T, Lucchesi C, Hostein I, Auzanneau C, Khalifa E, Soubeyran I, et al. High-depth sequencing of paired primary and metastatic tumours: implications for personalised medicine. Eur J Cancer. 2017; 84:250–6.
Article8. Manca A, Paliogiannis P, Colombino M, Casula M, Lissia A, Botti G, et al. Mutational concordance between primary and metastatic melanoma: a next-generation sequencing approach. J Transl Med. 2019; 17:289.
Article9. Crumley SM, Pepper KL, Phan AT, Olsen RJ, Schwartz MR, Portier BP. Next-generation sequencing of matched primary and metastatic rectal adenocarcinomas demonstrates minimal mutation gain and concordance to colonic adenocarcinomas. Arch Pathol Lab Med. 2016; 140:529–35.
Article10. McGranahan N, Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell. 2015; 27:15–26.
Article11. Cho M, Ahn S, Hong M, Bang H, Van Vrancken M, Kim S, et al. Tissue recommendations for precision cancer therapy using next generation sequencing: a comprehensive single cancer center’s experiences. Oncotarget. 2017; 8:42478–86.
Article12. Carithers LJ, Agarwal R, Guan P, Odeh H, Sachs MC, Engel KB, et al. The biospecimen preanalytical variables program: a multiassay comparison of effects of delay to fixation and fixation duration on nucleic acid quality. Arch Pathol Lab Med. 2019; 143:1106–18.
Article13. Chen G, Mosier S, Gocke CD, Lin MT, Eshleman JR. Cytosine deamination is a major cause of baseline noise in next-generation sequencing. Mol Diagn Ther. 2014; 18:587–93.
Article14. Kim S, Park C, Ji Y, Kim DG, Bae H, van Vrancken M, et al. Deamination effects in formalin-fixed, paraffin-embedded tissue samples in the era of precision medicine. J Mol Diagn. 2017; 19:137–46.
Article15. Ignatiadis M, Sledge GW, Jeffrey SS. Liquid biopsy enters the clinic: implementation issues and future challenges. Nat Rev Clin Oncol. 2021; 18:297–312.16. Mateo J, Chakravarty D, Dienstmann R, Jezdic S, Gonzalez-Perez A, Lopez-Bigas N, et al. A framework to rank genomic alterations as targets for cancer precision medicine: the ESMO Scale for Clinical Actionability of molecular Targets (ESCAT). Ann Oncol. 2018; 29:1895–902.
Article17. Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017; 2017:PO.1700011.
Article18. Jennings LJ, Arcila ME, Corless C, Kamel-Reid S, Lubin IM, Pfeifer J, et al. Guidelines for validation of next-generation sequencing-based oncology panels: a joint consensus recommendation of the Association for Molecular Pathology and College of American Pathologists. J Mol Diagn. 2017; 19:341–65.19. Heydt C, Wolwer CB, Velazquez Camacho O, Wagener-Ryczek S, Pappesch R, Siemanowski J, et al. Detection of gene fusions using targeted next-generation sequencing: a comparative evaluation. BMC Med Genomics. 2021; 14:62.
Article20. Strickler JH, Hanks BA, Khasraw M. Tumor mutational burden as a predictor of immunotherapy response: is more always better? Clin Cancer Res. 2021; 27:1236–41.
Article21. Marabelle A, Fakih M, Lopez J, Shah M, Shapira-Frommer R, Nakagawa K, et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 2020; 21:1353–65.
Article22. Fancello L, Gandini S, Pelicci PG, Mazzarella L. Tumor mutational burden quantification from targeted gene panels: major advancements and challenges. J Immunother Cancer. 2019; 7:183.
Article23. Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017; 357:409–13.24. Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015; 372:2509–20.25. Luchini C, Bibeau F, Ligtenberg MJ, Singh N, Nottegar A, Bosse T, et al. ESMO recommendations on microsatellite instability testing for immunotherapy in cancer, and its relationship with PD-1/PD-L1 expression and tumour mutational burden: a systematic review-based approach. Ann Oncol. 2019; 30:1232–43.
Article26. Dedeurwaerdere F, Claes KB, Van Dorpe J, Rottiers I, Van der Meulen J, Breyne J, et al. Comparison of microsatellite instability detection by immunohistochemistry and molecular techniques in colorectal and endometrial cancer. Sci Rep. 2021; 11:12880.
Article27. Luchini C, Lawlor RT, Milella M, Scarpa A. Molecular tumor boards in clinical practice. Trends Cancer. 2020; 6:738–44.
Article28. van der Velden DL, van Herpen CM, van Laarhoven HW, Smit EF, Groen HJ, Willems SM, et al. Molecular tumor boards: current practice and future needs. Ann Oncol. 2017; 28:3070–5.
Article29. Larson KL, Huang B, Weiss HL, Hull P, Westgate PM, Miller RW, et al. Clinical outcomes of molecular tumor boards: a systematic review. JCO Precis Oncol. 2021; 5:PO.20.00495.
Article30. Pishvaian MJ, Blais EM, Bender RJ, Rao S, Boca SM, Chung V, et al. A virtual molecular tumor board to improve efficiency and scalability of delivering precision oncology to physicians and their patients. JAMIA Open. 2019; 2:505–15.
Article31. Rao S, Pitel B, Wagner AH, Boca SM, McCoy M, King I, et al. Collaborative, multidisciplinary evaluation of cancer variants through virtual molecular tumor boards informs local clinical practices. JCO Clin Cancer Inform. 2020; 4:602–13.
Article32. Health Insurance Review and Assessment Service. Process for off-label drug use for chemotherapy [Internet]. Wonju: Health Insurance Review and Assessment Service;2018. [cited 2021 Dec 8]. Available from: https://www.hira.or.kr/bbsDummy.do?pgmid=HIRAA030023080000&brdScnBltNo=4&brdBltNo=45637 .33. Clinical Trial Policy Division, Pharmaceutical Safety Bureau, Ministry of Food and Drug Safety. Guideline for expanded access program [Internet]. Cheongju: Ministry of Food and Drug Safety;2020. [cited 2021 Dec 8]. Available from: https://www.mfds.go.kr/brd/m_1060/view.do?seq=14576&srchFr=&srchTo=&srchWord=&srchTp=&itm_seq_1=0&itm_seq_2=0&multi_itm_seq=0&company_cd=&company_nm=&page=22 .34. Walsh S, de Jong EE, van Timmeren JE, Ibrahim A, Compter I, Peerlings J, et al. Decision support systems in oncology. JCO Clin Cancer Inform. 2019; 3:1–9.
Article35. Blasi L, Bordonaro R, Serretta V, Piazza D, Firenze A, Gebbia V. Virtual clinical and precision medicine tumor boards-cloud-based platform-mediated implementation of multidisciplinary reviews among oncology centers in the COVID-19 era: protocol for an observational study. JMIR Res Protoc. 2021; 10:e26220.
Article36. Kato S, Kim KH, Lim HJ, Boichard A, Nikanjam M, Weihe E, et al. Real-world data from a molecular tumor board demonstrates improved outcomes with a precision N-of-One strategy. Nat Commun. 2020; 11:4965.
Article37. Choi YJ, Choi JY, Kim JW, Lim AR, Lee Y, Chang WJ, et al. Comparison of the data of a next-generation sequencing panel from K-MASTER project with that of orthogonal methods for detecting targetable genetic alterations. Cancer Res Treat. 2021. May. 20. [Epub]. https://doi.org/10.4143/crt.2021.218 .
Article38. Nakamura Y, Fujisawa T, Taniguchi H, Bando H, Okamoto W, Tsuchihara K, et al. SCRUM-Japan GI-SCREEN and MONSTAR-SCREEN: path to the realization of biomarker-guided precision oncology in advanced solid tumors. Cancer Sci. 2021; 112:4425–32.
Article39. Barlesi F, Mazieres J, Merlio JP, Debieuvre D, Mosser J, Lena H, et al. Routine molecular profiling of patients with advanced non-small-cell lung cancer: results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT). Lancet. 2016; 387:1415–26.
Article40. Lee Y, Lee S, Sung JS, Chung HJ, Lim AR, Kim JW, et al. Clinical application of targeted deep sequencing in metastatic colorectal cancer patients: actionable genomic alteration in K-MASTER project. Cancer Res Treat. 2021; 53:123–30.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- ERRATUM: Recommendations for the Use of Next-Generation Sequencing and the Molecular Tumor Board for Patients with Advanced Cancer: A Report from KSMO and KCSG Precision Medicine Networking Group
- Clinical Implication of Molecular Tumor Board
- Latest trends in cancer clinical trials using genomics
- Ultra-rare Disease and Genomics-Driven Precision Medicine
- The Cancer Precision Medicine Diagnosis and Treatment (K-MASTER) Enterprise