Cancer Res Treat.
2021 Oct;53(4):944-961. 10.4143/crt.2020.466.
Correlation of NUF2 Overexpression with Poorer Patient Survival in Multiple Cancers
- Affiliations
-
- 1Department of Otorhinolaryngology-Head and Neck Surgery, Key Laboratory, The Affiliated Hospital of Qingdao University, Qingdao, China
- 2Department of Oncological Surgery, Zhejiang Shangyu People's Hospital, Shaoxing, China
- 3Division of Cellular and Molecular Research, National Cancer Centre Singapore, Singapore
- 4Department of Biological Specimen Bank, The Affiliated Hospital of Qingdao University, Qingdao, China
- 5Department of Pathology, The Affiliated Hospital of Qingdao University, Qingdao, China
- 6Department of Hepato-Pancreato-Biliary Surgery, Singapore General Hospital, Singapore
- 7Department of Biochemistry, Yong Loo Lin School of Medicine, National University of Singapore, Singapore
- 8Institute of Molecular and Cell Biology, A*STAR, Singapore
- 9Duke-NUS Medical School, Singapore
- 10School of Medicine, Hangzhou Normal University, Hangzhou, China
- KMID: 2521571
- DOI: http://doi.org/10.4143/crt.2020.466
Abstract
- Purpose
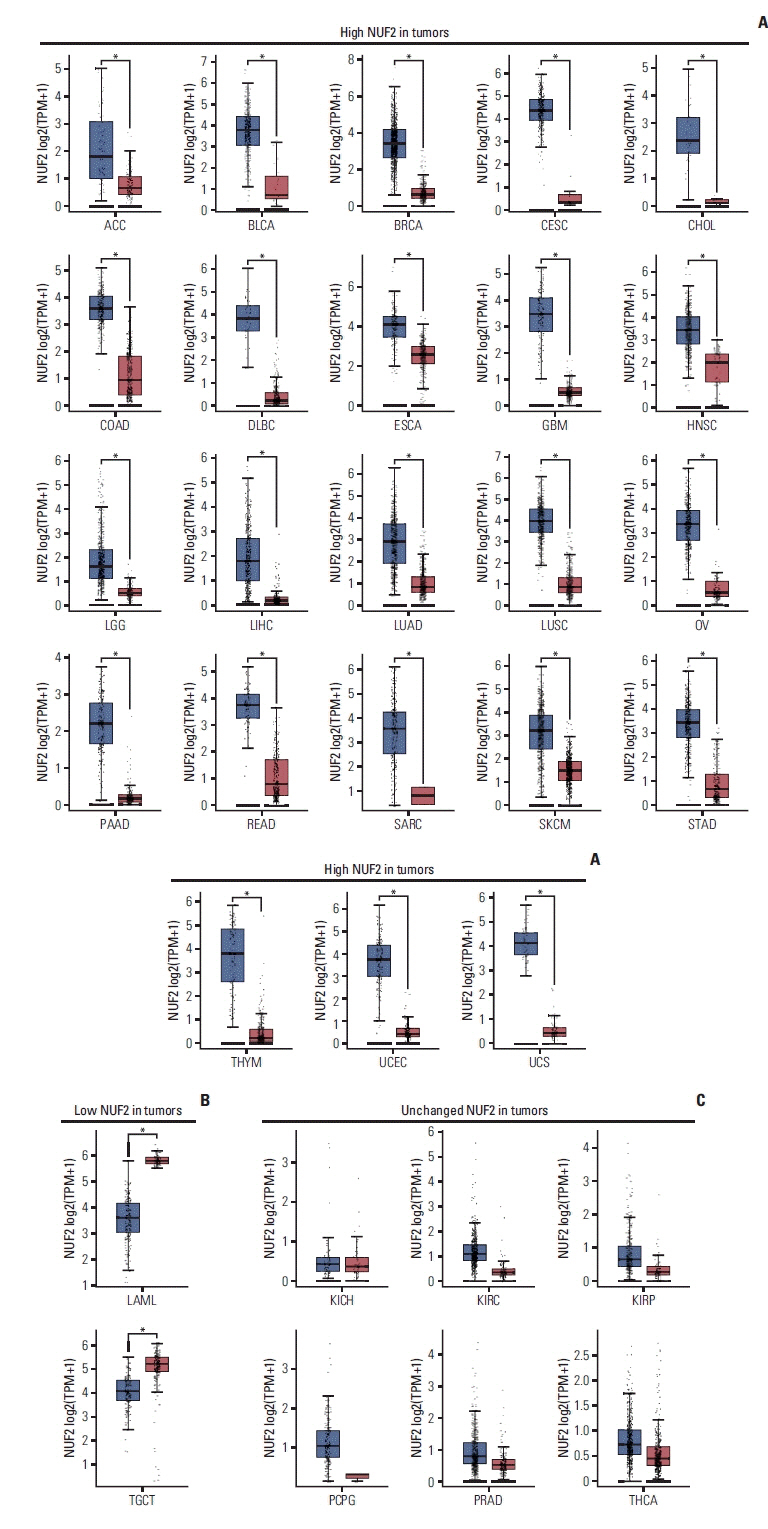
NUF2 has been implicated in multiple cancers recently, suggesting NUF2 may play a role in the common tumorigenesis process. In this study, we aim to perform comprehensive meta-analysis of NUF2 expression in the cancer types included in the Cancer Genome Atlas (TCGA).
Materials and Methods
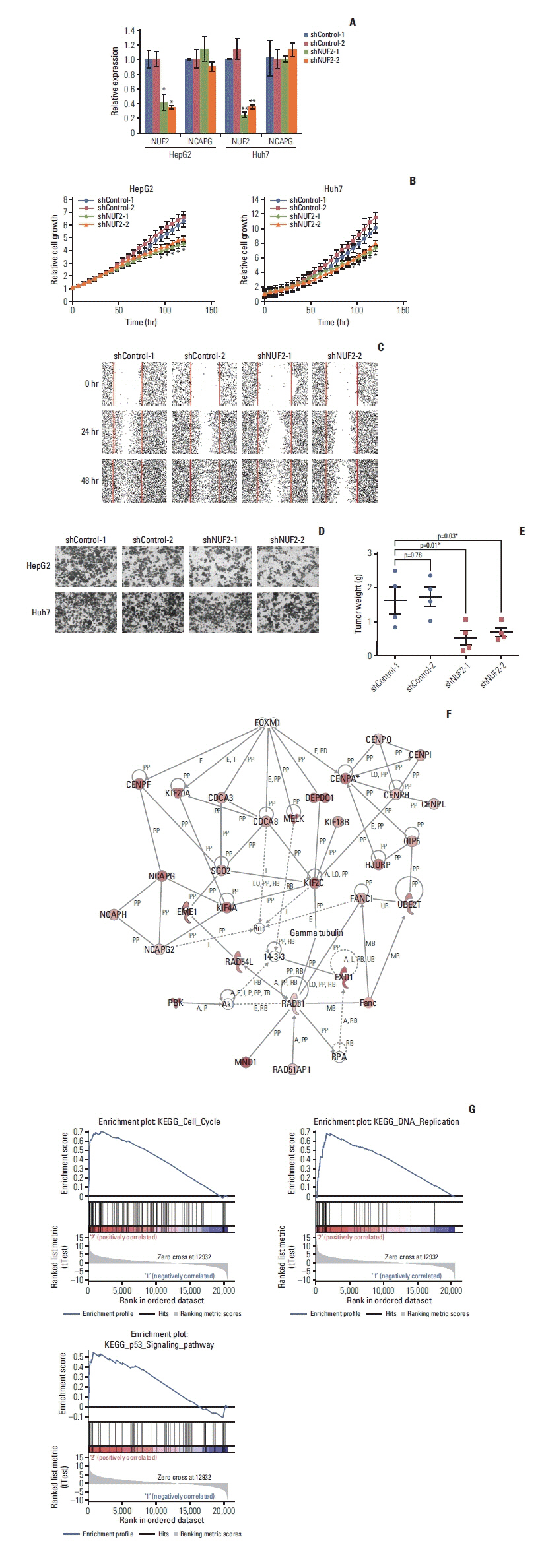
RNA-sequencing data in 31 cancer types in the TCGA data and 11 independent datasets were used to examine NUF2 expression. Silencing NUF2 using targeting shRNAs in hepatocellular carcinoma (HCC) cell lines was used to evaluate NUF2’s role in HCC in vitro and in vivo.
Results
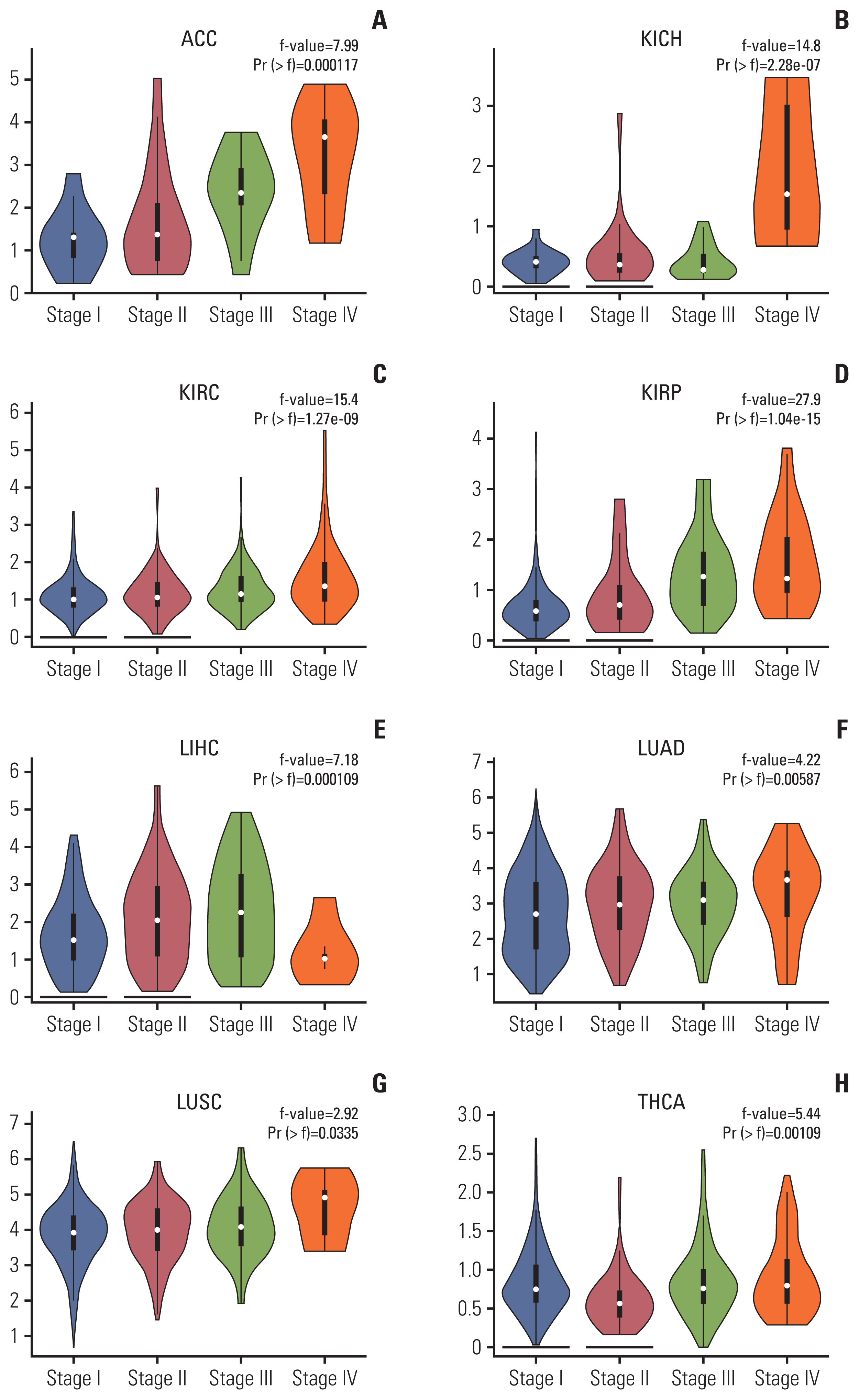
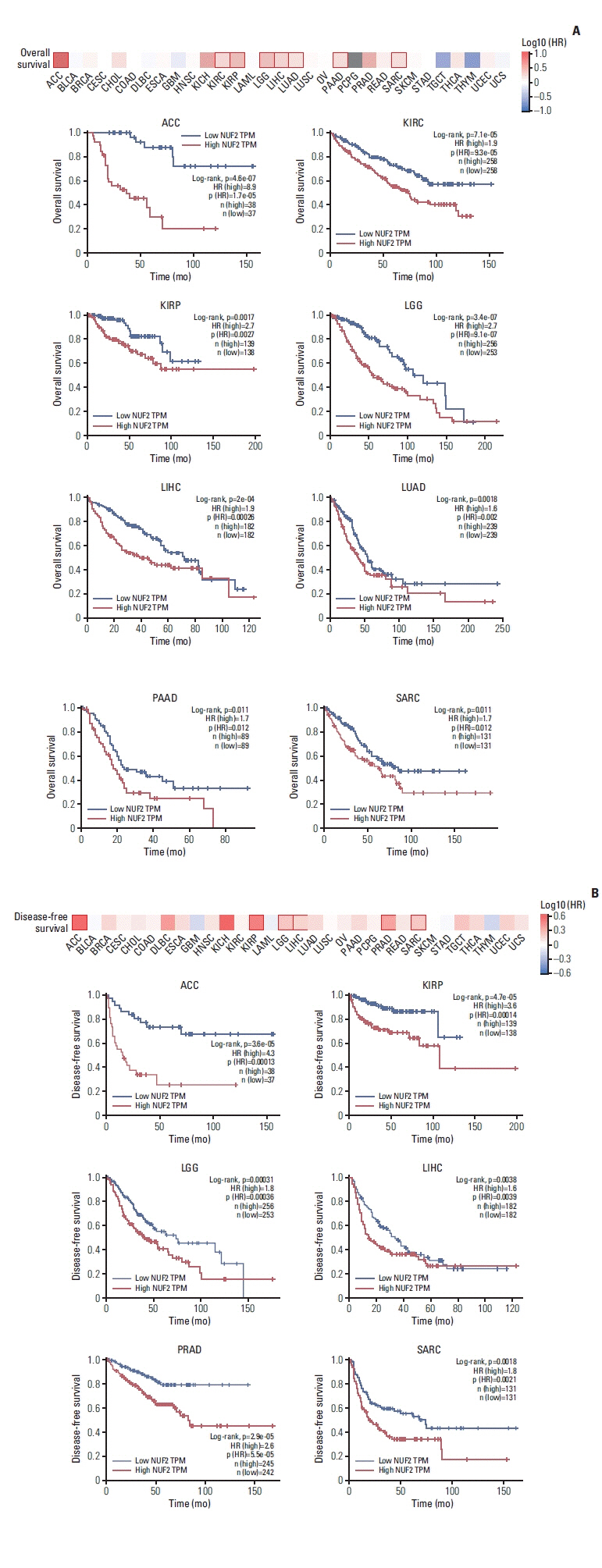
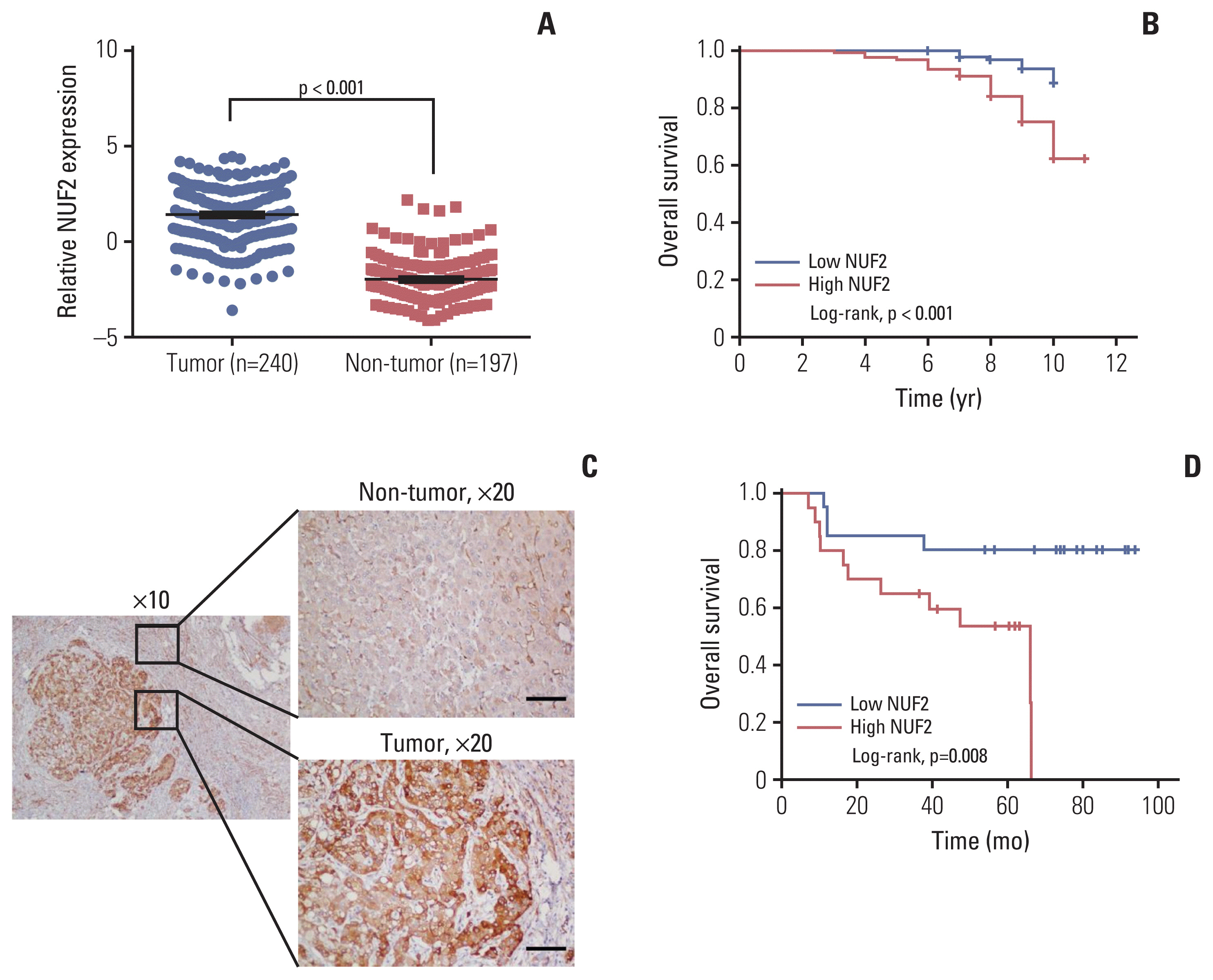
NUF2 up-regulation is significantly observed in 23 out of the 31 cancer types in the TCGA datasets and validated in 13 major cancer types using 11 independent datasets. NUF2 overexpression was clinically important as high NUF2 was significantly associated with tumor stages in eight different cancers. High NUF2 was also associated with significantly poorer patient overall survival and disease-free survival in eight and six cancers, respectively. We proceeded to validate NUF2 overexpression and its negative association with overall survival at the protein level in an independent cohort of 40 HCC patients. Compared to the non-targeting controls, NUF2 knockdown cells showed significantly reduced ability to grow, migrate into a scratch wound and invade the 8 μm porous membrane in vitro. Moreover, NUF2 knockdown cells also formed significantly smaller tumors than control cells in mouse xenograft assays in vivo.
Conclusion
NUF2 up-regulation is a common feature of many cancers. The prognostic potential and functional impact of NUF2 up-regulation warrant further studies.
Keyword
Figure
Reference
-
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018; 68:394–424.
Article2. Stewart BW, Wild CP. World cancer report 2014. Lyon: International Agency for Research on Cancer;2014.3. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011; 144:646–74.
Article4. Nabetani A, Koujin T, Tsutsumi C, Haraguchi T, Hiraoka Y. A conserved protein, Nuf2, is implicated in connecting the centromere to the spindle during chromosome segregation: a link between the kinetochore function and the spindle checkpoint. Chromosoma. 2001; 110:322–34.
Article5. Wang Y, Tan PY, Handoko YA, Sekar K, Shi M, Xie C, et al. NUF2 is a valuable prognostic biomarker to predict early recurrence of hepatocellular carcinoma after surgical resection. Int J Cancer. 2019; 145:662–70.
Article6. Xu W, Wang Y, Wang Y, Lv S, Xu X, Dong X. Screening of differentially expressed genes and identification of NUF2 as a prognostic marker in breast cancer. Int J Mol Med. 2019; 44:390–404.
Article7. Huang SK, Qian JX, Yuan BQ, Lin YY, Ye ZX, Huang SS. SiRNA-mediated knockdown against NUF2 suppresses tumor growth and induces cell apoptosis in human glioma cells. Cell Mol Biol (Noisy-le-grand). 2014; 60:30–6.8. Sugimasa H, Taniue K, Kurimoto A, Takeda Y, Kawasaki Y, Akiyama T. Heterogeneous nuclear ribonucleoprotein K upregulates the kinetochore complex component NUF2 and promotes the tumorigenicity of colon cancer cells. Biochem Biophys Res Commun. 2015; 459:29–35.
Article9. Liu Q, Dai SJ, Li H, Dong L, Peng YP. Silencing of NUF2 inhibits tumor growth and induces apoptosis in human hepatocellular carcinomas. Asian Pac J Cancer Prev. 2014; 15:8623–9.
Article10. Hu P, Chen X, Sun J, Bie P, Zhang LD. siRNA-mediated knockdown against NUF2 suppresses pancreatic cancer proliferation in vitro and in vivo. Biosci Rep. 2015; 35:e00170.
Article11. Hu P, Shangguan J, Zhang L. Downregulation of NUF2 inhibits tumor growth and induces apoptosis by regulating lncRNA AF339813. Int J Clin Exp Pathol. 2015; 8:2638–48.12. Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016; 534:47–54.13. Blum A, Wang P, Zenklusen JC. SnapShot: TCGA-analyzed tumors. Cell. 2018; 173:530.
Article14. Hutter C, Zenklusen JC. The Cancer Genome Atlas: creating lasting value beyond its data. Cell. 2018; 173:283–5.
Article15. Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019; 47:W556–60.
Article16. Wang Y, Gao B, Tan PY, Handoko YA, Sekar K, Deivasigamani A, et al. Genome-wide CRISPR knockout screens identify NCAPG as an essential oncogene for hepatocellular carcinoma tumor growth. FASEB J. 2019; 33:8759–70.
Article17. Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003; 34:267–73.
Article18. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005; 102:15545–50.
Article19. Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014; 158:929–44.
Article20. Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012; 483:603–7.21. Ghandi M, Huang FW, Jane-Valbuena J, Kryukov GV, Lo CC, McDonald ER 3rd, et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019; 569:503–8.22. DeLuca JG, Moree B, Hickey JM, Kilmartin JV, Salmon ED. hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J Cell Biol. 2002; 159:549–55.
Article23. Liu D, Ding X, Du J, Cai X, Huang Y, Ward T, et al. Human NUF2 interacts with centromere-associated protein E and is essential for a stable spindle microtubule-kinetochore attachment. J Biol Chem. 2007; 282:21415–24.
Article24. Sundin LJ, Guimaraes GJ, Deluca JG. The NDC80 complex proteins Nuf2 and Hec1 make distinct contributions to kinetochore-microtubule attachment in mitosis. Mol Biol Cell. 2011; 22:759–68.
Article25. Gu L, Zhang L, Hou N, Li M, Shen W, Xie X, et al. Clinical and radiographic characterization of primary seminomas and nonseminomatous germ cell tumors. Niger J Clin Pract. 2019; 22:342–9.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Prognostic Significance of p53 Overexpression after Hepatic Resection of Hepatocellular Carcinoma
- The alteration of p16 protein in invasive cervical cancer
- The expression of clusterin, bax, p53, Ki-67, and apoptotic index in epithelial ovarian tumors
- Amplification and Overexpression of c-erbB2/neu in Bladder Tumor Using Real-time Quantitative PCR with TaqMan Detection System
- Amplification and Overexpression of Cyclin D1 in Head and Neck Squamous Cell Carcinomas