Healthc Inform Res.
2021 Jul;27(3):189-199. 10.4258/hir.2021.27.3.189.
Impact of the Choice of Cross-Validation Techniques on the Results of Machine Learning-Based Diagnostic Applications
- Affiliations
-
- 1Electronic Systems Sensors and Nanobiotechnologies (E2SN), ENSAM, Mohammed V University in Rabat, Morocco
- KMID: 2519035
- DOI: http://doi.org/10.4258/hir.2021.27.3.189
Abstract
Objectives
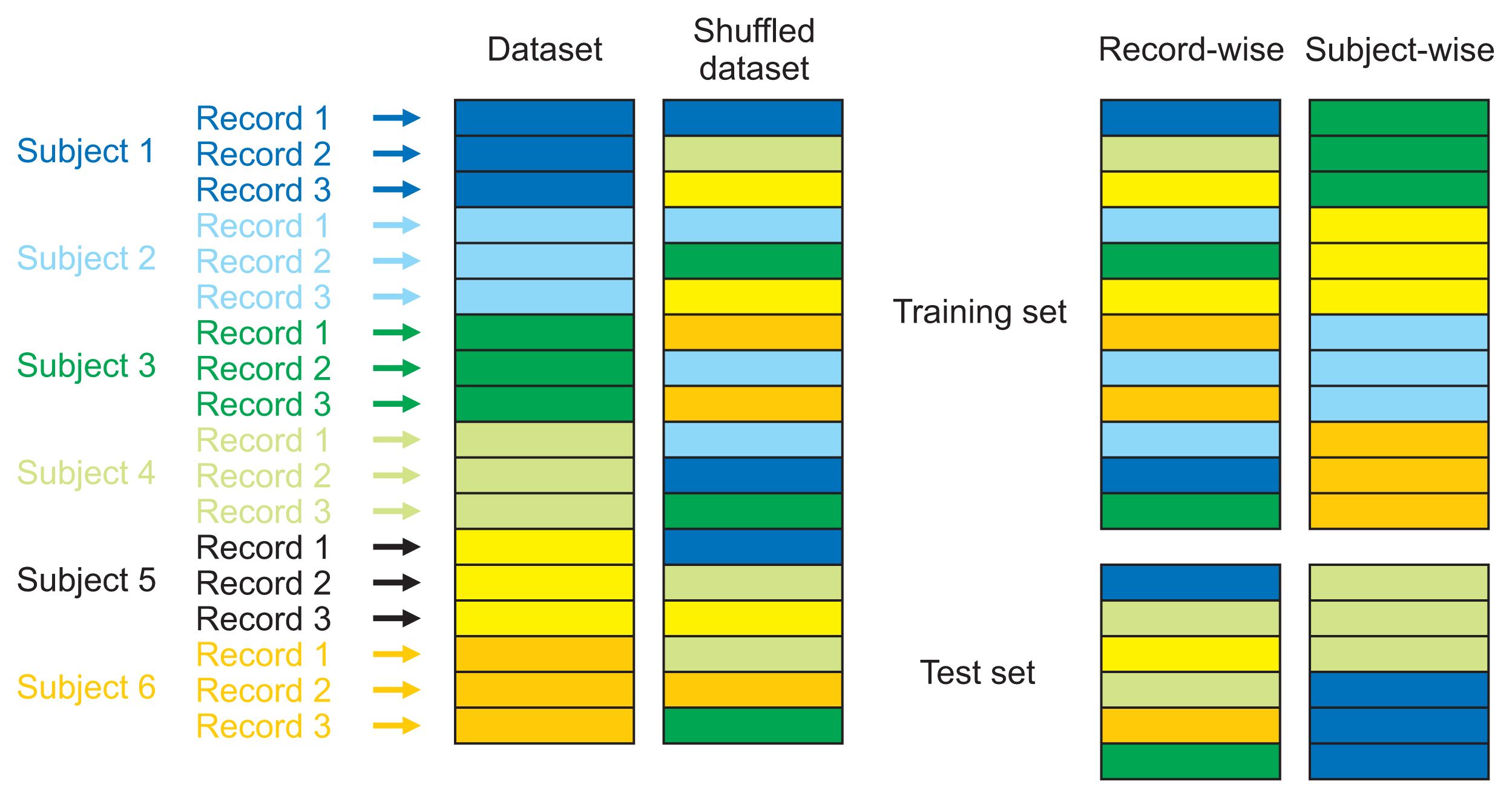
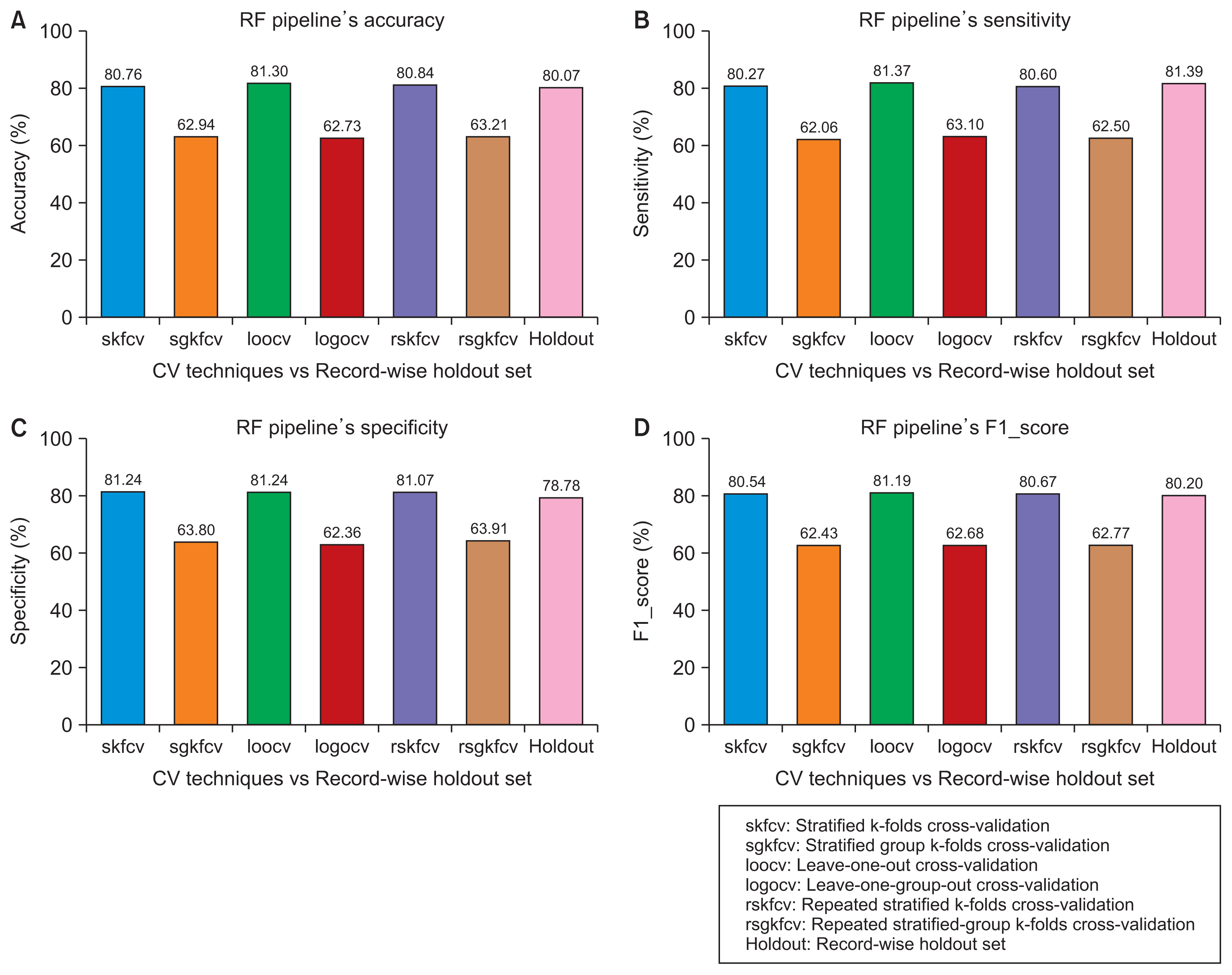
With advances in data availability and computing capabilities, artificial intelligence and machine learning technologies have evolved rapidly in recent years. Researchers have taken advantage of these developments in healthcare informatics and created reliable tools to predict or classify diseases using machine learning-based algorithms. To correctly quantify the performance of those algorithms, the standard approach is to use cross-validation, where the algorithm is trained on a training set, and its performance is measured on a validation set. Both datasets should be subject-independent to simulate the expected behavior of a clinical study. This study compares two cross-validation strategies, the subject-wise and the record-wise techniques; the subject-wise strategy correctly mimics the process of a clinical study, while the record-wise strategy does not.
Methods
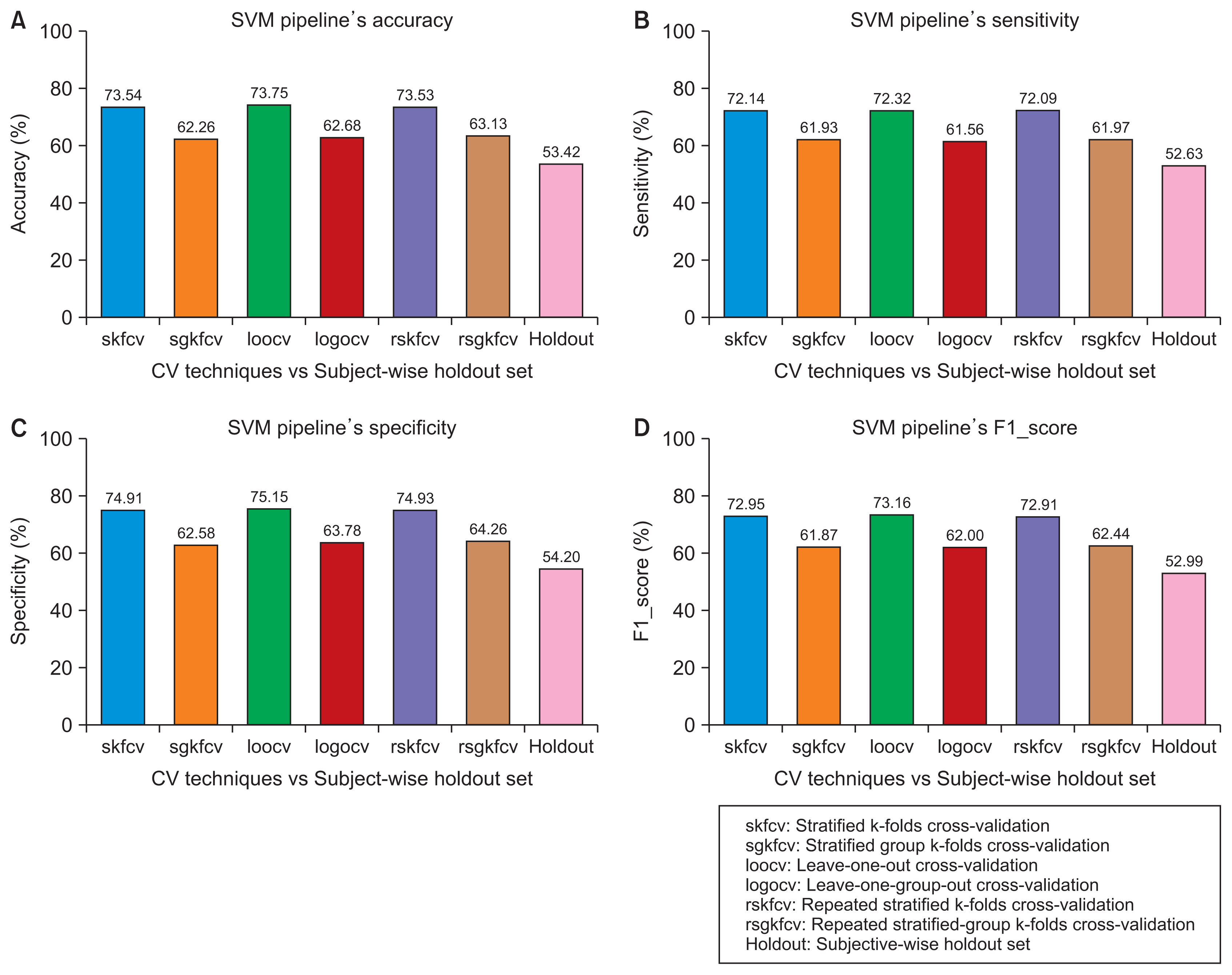
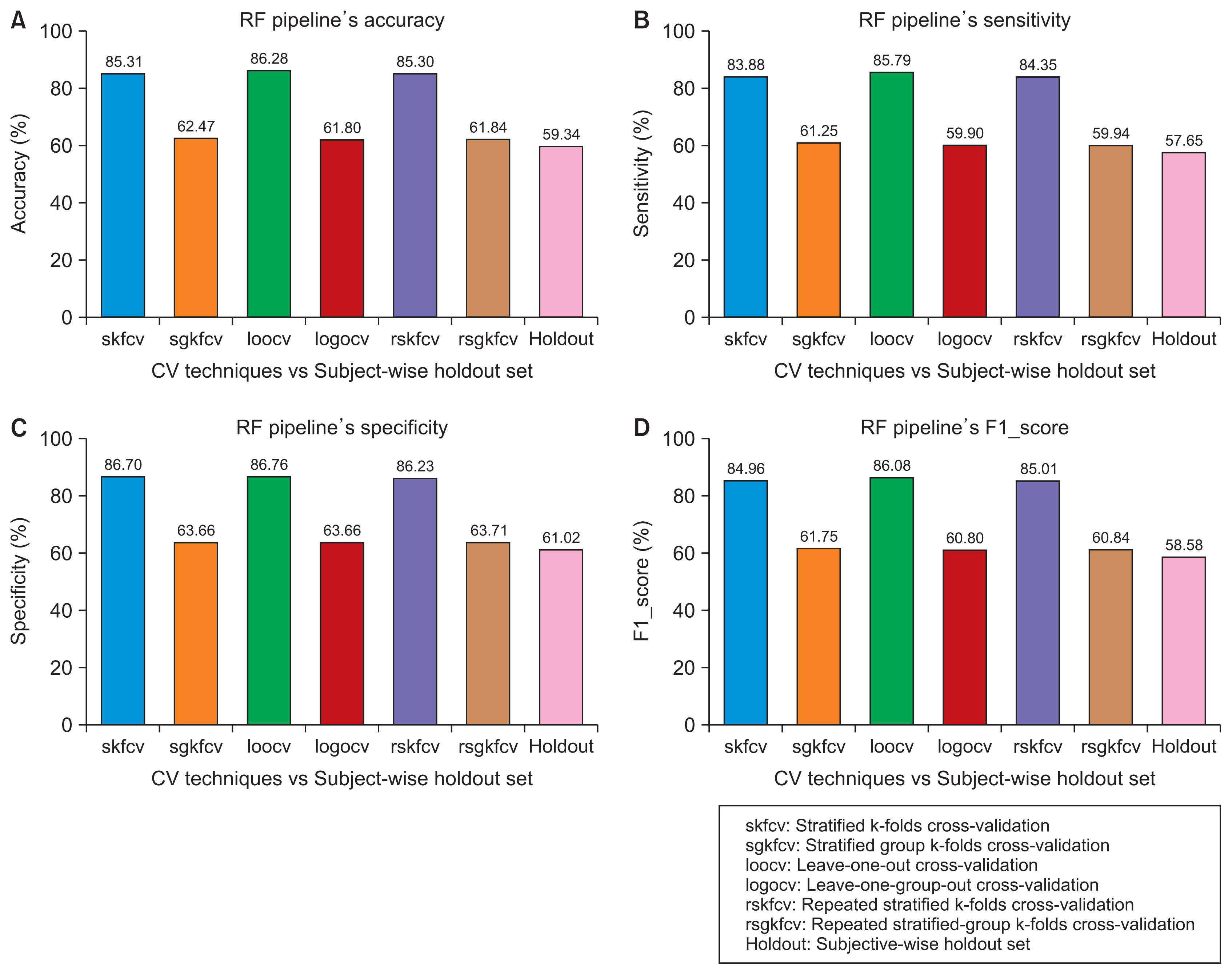
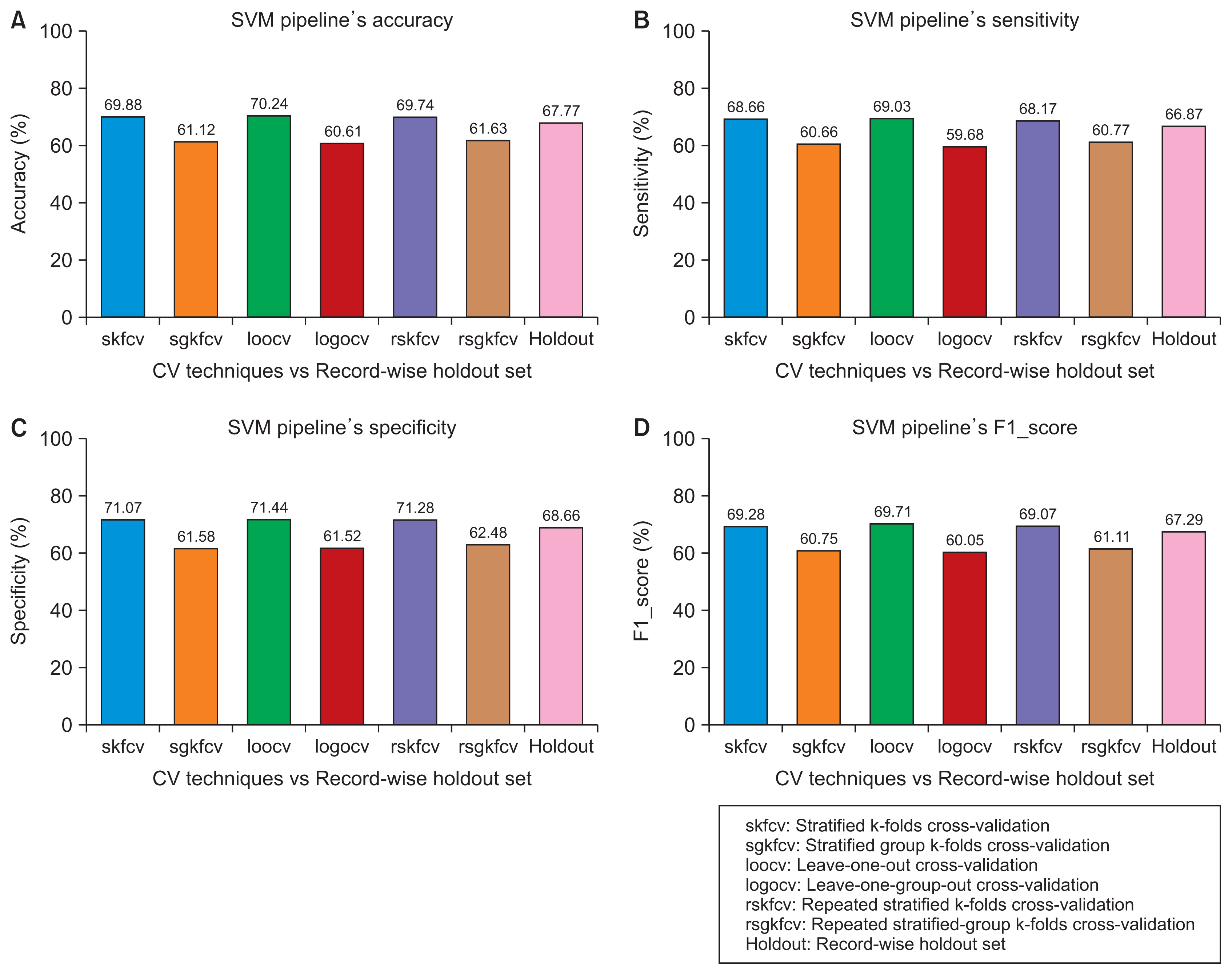
We started by creating a dataset of smartphone audio recordings of subjects diagnosed with and without Parkinson’s disease. This dataset was then divided into training and holdout sets using subject-wise and the record-wise divisions. The training set was used to measure the performance of two classifiers (support vector machine and random forest) to compare six cross-validation techniques that simulated either the subject-wise process or the record-wise process. The holdout set was used to calculate the true error of the classifiers.
Results
The record-wise division and the record-wise cross-validation techniques overestimated the performance of the classifiers and underestimated the classification error.
Conclusions
In a diagnostic scenario, the subject-wise technique is the proper way of estimating a model’s performance, and record-wise techniques should be avoided.
Figure
Reference
-
References
1. Perry B, Herrington W, Goldsack JC, Grandinetti CA, Vasisht KP, Landray MJ, et al. Use of mobile devices to measure outcomes in clinical research, 2010–2016: a systematic literature review. Digit Biomark. 2018; 2(1):11–30.
Article2. Arlot S, Celisse A. A survey of cross-validation procedures for model selection. Stat Surv. 2010; 4:40–79.
Article3. Bot BM, Suver C, Neto EC, Kellen M, Klein A, Bare C, et al. The mPower study, Parkinson disease mobile data collected using ResearchKit. Sci Data. 2016; 3:160011.
Article4. Sage Bionetworks. The mPower Public Researcher Portal [Internet]. Seattle (WA): Sage Bionetworks;2019. [cited at 2021 Aug 3]. Available from: https://www.synapse.org/#!Synapse:syn4993293/wiki/247859 .5. Sage Bionetworks. Synapse REST API: Basics Table Query [Internet]. Seattle (WA): Sage Bionetworks;2016. [cited at 2021 Aug 3]. Available from: https://docs.synapse.org/rest/org/sagebionetworks/repo/web/controller/TableExamples.html .6. Rose S, Laan MJ. Why match? Investigating matched case-control study designs with causal effect estimation. Int J Biostat. 2009; 5(1):1.
Article7. Wong SL, Gilmour H, Ramage-Morin PL. Parkinson’s disease: prevalence, diagnosis and impact. Health Rep. 2014; 25(11):10–4.8. Khalid S, Khalil T, Nasreen S. A survey of feature selection and feature extraction techniques in machine learning. In : Proceedings of 2014 Science and Information Conference; 2014 Aug 27–29; London, UK. p. 372–8.
Article9. Giannakopoulos T, Pikrakis A. Introduction to audio analysis: a MATLAB approach. San Diego, CA: Academic Press;2014.10. Giannakopoulos T. pyAudioAnalysis: an open-source python library for audio signal analysis. PLoS One. 2015; 10(12):e0144610.
Article11. Hawkins DM, Basak SC, Mills D. Assessing model fit by cross-validation. J Chem Inf Comput Sci. 2003; 43(2):579–86.
Article12. Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011; 12:2825–30.13. Github. Stratified GroupKFold [Internet]. San Francisco (CA): Github.com;2019. [cited at 2021 Aug 3]. Available from: https://github.com/scikit-learn/scikit-learn/issues/13621 .14. Kotsiantis SB, Kanellopoulos D, Pintelas PE. Data preprocessing for supervised leaning. Int J Comput Sci. 2006; 1(2):111–7.15. Chandrashekar G, Sahin F. A survey on feature selection methods. Comput Electr Eng. 2014; 40(1):16–28.
Article16. Zou H, Hastie T. Regularization and variable selection via the elastic net. J R Stat Soc Series B Stat Methodol. 2005; 67(2):301–20.
Article17. Wager S. Cross-validation, risk estimation, and model selection: comment on a paper by Rosset and Tibshirani. J Am Stat Assoc. 2020; 115(529):157–60.
Article18. Saeb S, Lonini L, Jayaraman A, Mohr DC, Kording KP. The need to approximate the use-case in clinical machine learning. Gigascience. 2017; 6(5):1–9.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Application of Machine Learning in Rhinology: A State of the Art Review
- Applications of Machine Learning Using Electronic Medical Records in Spine Surgery
- Diagnostic Accuracy of Machine Learning Algorithms for Hepatitis A Antibody
- Machine Learning Smart System for Parkinson Disease Classification Using the Voice as a Biomarker
- Simultaneous Utilization of Mood Disorder Questionnaire and Bipolar Spectrum Diagnostic Scale for Machine Learning-Based Classification of Patients With Bipolar Disorders and Depressive Disorders