Korean J Transplant.
2021 Mar;35(1):1-7. 10.4285/kjt.20.0035.
Comparison of single nucleotide polymorphisms and short tandem repeats as markers for differentiating between donors and recipients in solid organ transplantation
- Affiliations
-
- 1Department of Laboratory Medicine, and Research Institute for Convergence of Biomedical Science and Technology, Pusan National University Yangsan Hospital, Yangsan, Korea
- 2Department of Laboratory Medicine, Pusan National University Hospital, Busan, Korea
- KMID: 2514402
- DOI: http://doi.org/10.4285/kjt.20.0035
Abstract
- Background
To analyze transplant rejection and to distinguish between donor and recipient, it is necessary to select a marker from single nucleotide polymorphism (SNP), short tandem repeat (STR), and human leukocyte antigen (HLA) testing. SNPs are bi-allelic and the polymerase chain reaction method used for SNP testing has the advantage of lower cost than sequencing methods. In this study, we aimed to distinguish donors from recipients using a combination of existing commercialized STRs and the SNPs identified.
Methods
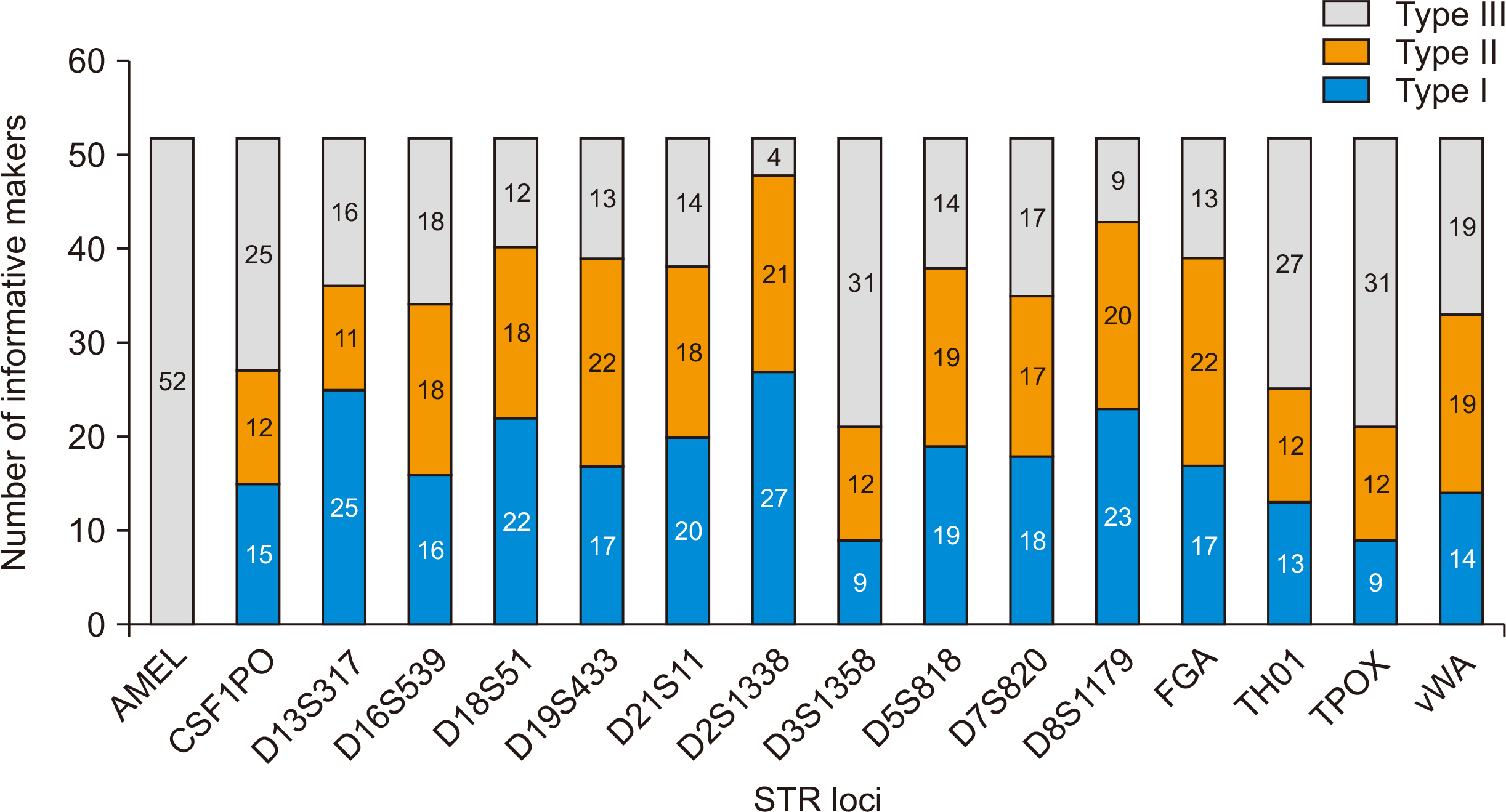
All selected SNPs complied with the following criterion: known and validated minor allele frequency (MAF) ≥43% in Korean and reported ethnicities from global pop-ulations (HapMap, 1000 Genomes, and the Korean Reference Genome project). The STR assays were performed for 16 tetranucleotide repeat loci.
Results
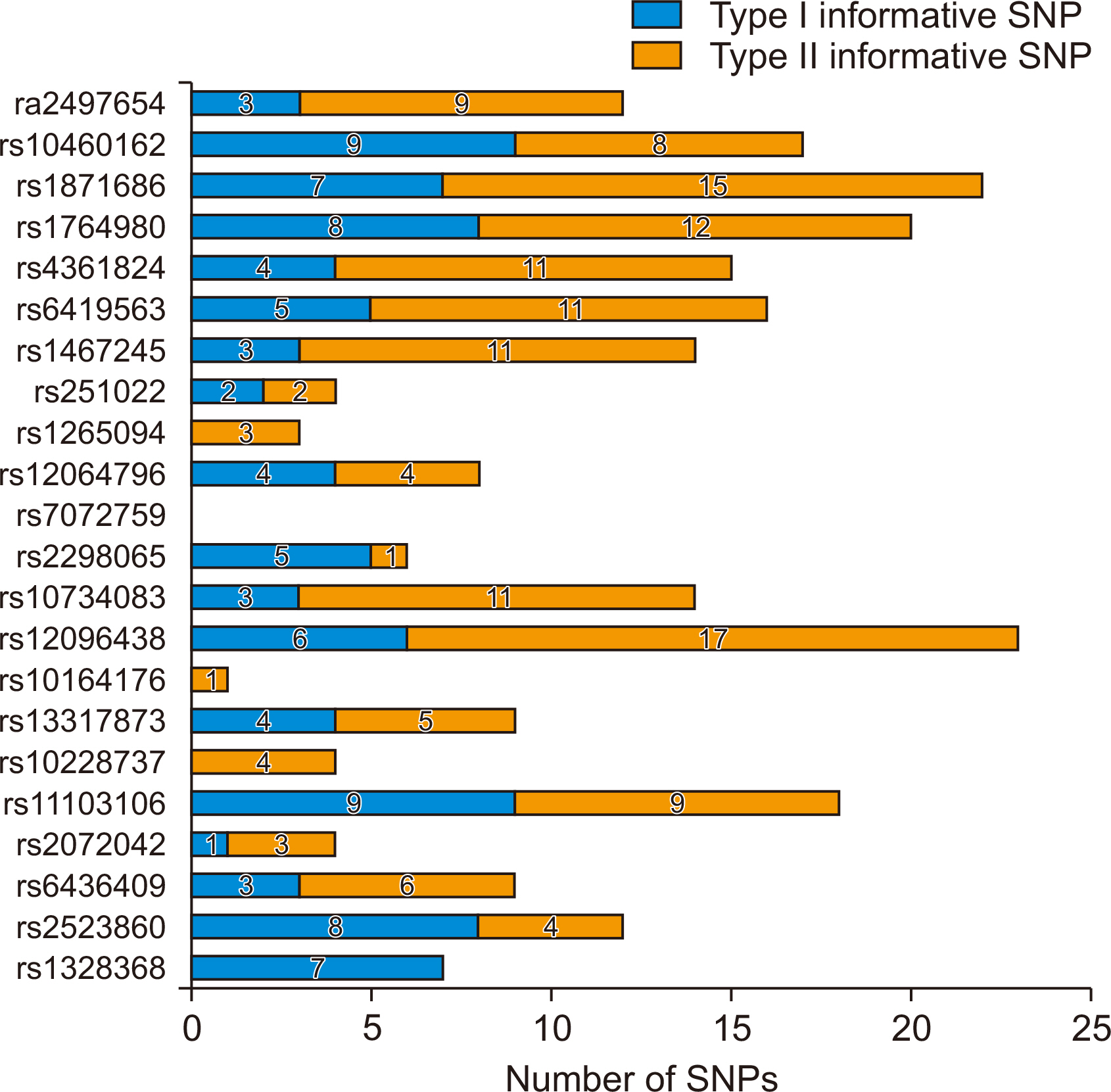
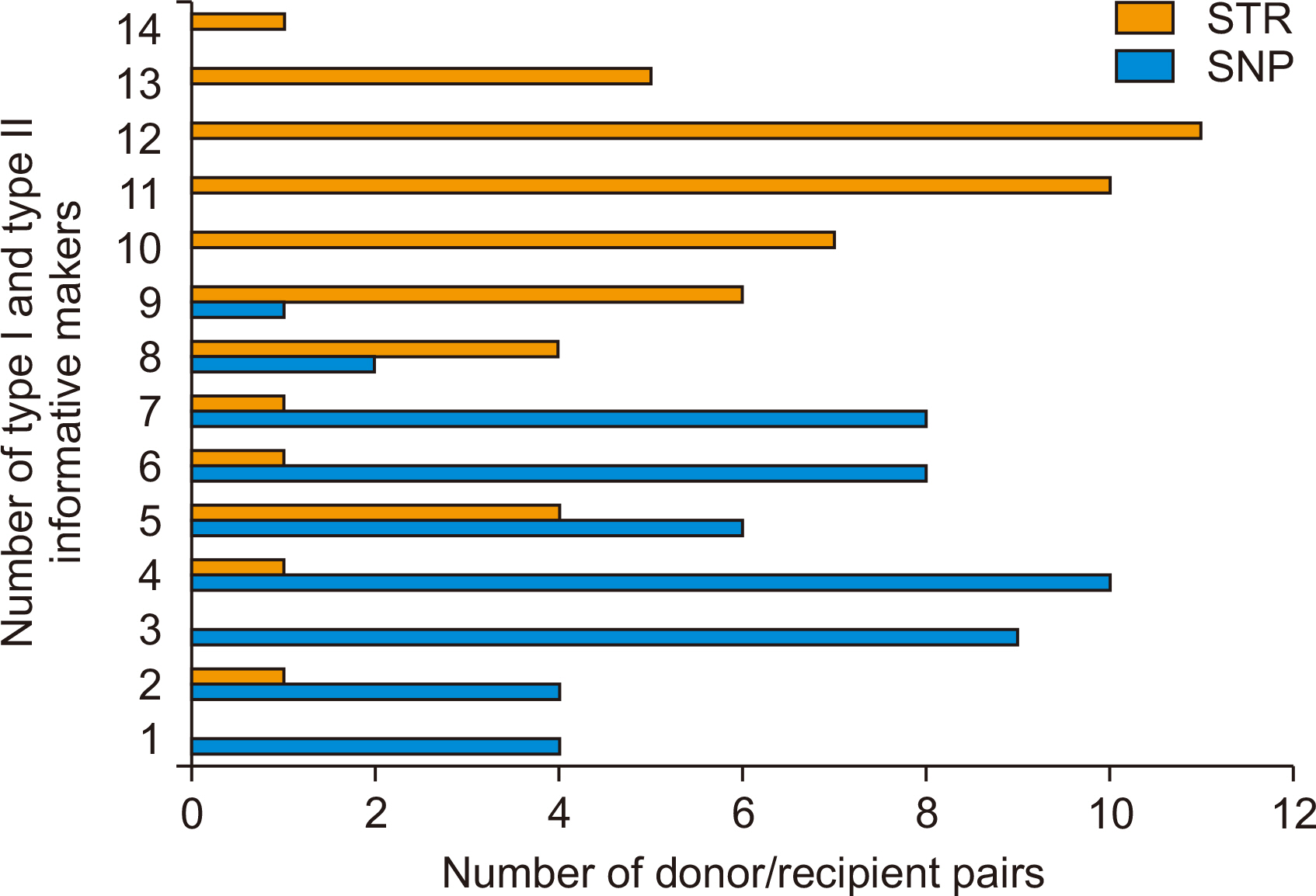
DNA from the 52 donor/recipient pairs were tested for informative markers. The median age of the recipients was 47 years. MAF in the 52 pairs was 1.0%–76.0%. The probability of informative genotypes (I) was 0.001–0.124. The summation of I was 0.680. In the 52 donor recipient pairs, the selected SNPs showed a 0.031 average probability of being informative. The probability of identity in our study was 0.122–0.348. SNP panel configuration distinguished 100% of 52 donors/recipient pairs.
Conclusions
Donors and recipients were distinguished by STR and 22 SNPs with MAF identified from SNP databases. Seventeen SNPs were able to distinguish between donors and recipients (I value=0.039).
Figure
Reference
-
1. Snyder TM, Khush KK, Valantine HA, Quake SR. 2011; Universal noninvasive detection of solid organ transplant rejection. Proc Natl Acad Sci U S A. 108:6229–34. DOI: 10.1073/pnas.1013924108. PMID: 21444804. PMCID: PMC3076856.
Article2. Gielis EM, Ledeganck KJ, De Winter BY, Del Favero J, Bosmans JL, Claas FH, et al. 2015; Cell-free DNA: an upcoming biomarker in transplantation. Am J Transplant. 15:2541–51. DOI: 10.1111/ajt.13387. PMID: 26184824.
Article3. Pontes ML, Fondevila M, Laréu MV, Medeiros R. 2015; SNP markers as additional information to resolve complex kinship cases. Transfus Med Hemother. 42:385–8. DOI: 10.1159/000440832. PMID: 26733770. PMCID: PMC4698646.
Article4. Beck J, Bierau S, Balzer S, Andag R, Kanzow P, Schmitz J, et al. 2013; Digital droplet PCR for rapid quantification of donor DNA in the circulation of transplant recipients as a potential universal biomarker of graft injury. Clin Chem. 59:1732–41. DOI: 10.1373/clinchem.2013.210328. PMID: 24061615.
Article5. Thongprayoon C, Vaitla P, Craici IM, Leeaphorn N, Hansrivijit P, Salim SA, et al. 2020; The use of donor-derived cell-free DNA for assessment of allograft rejection and injury status. J Clin Med. 9:1480. DOI: 10.3390/jcm9051480. PMID: 32423115. PMCID: PMC7290747.
Article6. Knight SR, Thorne A, Lo Faro ML. 2019; Donor-specific cell-free DNA as a biomarker in solid organ transplantation: a systematic review. Transplantation. 103:273–83. DOI: 10.1097/TP.0000000000002482. PMID: 30308576.
Article7. Li L, Wang Y, Yang S, Xia M, Yang Y, Wang J, et al. 2017; Genome-wide screening for highly discriminative SNPs for personal identification and their assessment in world populations. Forensic Sci Int Genet. 28:118–27. DOI: 10.1016/j.fsigen.2017.02.005. PMID: 28249201.
Article8. Børsting C, Morling N. Kumar D, Antonarakis S, editors. 2016. Genomic applications in forensic medicine. Medical and Health Genomics. Academic Press;Cambridge, MA: p. 295–309. DOI: 10.1016/B978-0-12-420196-5.00022-8.
Article9. Shiina T, Hosomichi K, Inoko H, Kulski JK. 2009; The HLA genomic loci map: expression, interaction, diversity and disease. J Hum Genet. 54:15–39. DOI: 10.1038/jhg.2008.5. PMID: 19158813.
Article10. Castella V, Gervaix J, Hall D. 2013; DIP-STR: highly sensitive markers for the analysis of unbalanced genomic mixtures. Hum Mutat. 34:644–54. DOI: 10.1002/humu.22280. PMID: 23355272. PMCID: PMC3675636.11. Oldoni F, Podini D. 2019; Forensic molecular biomarkers for mixture analysis. Forensic Sci Int Genet. 41:107–19. DOI: 10.1016/j.fsigen.2019.04.003. PMID: 31071519.
Article12. Robin JD, Ludlow AT, LaRanger R, Wright WE, Shay JW. 2016; Comparison of DNA quantification methods for next generation sequencing. Sci Rep. 6:24067. DOI: 10.1038/srep24067. PMID: 27048884. PMCID: PMC4822169.
Article13. Quan PL, Sauzade M, Brouzes E. 2018; dPCR: a technology review. Sensors (Basel). 18:1271. DOI: 10.3390/s18041271. PMID: 29677144. PMCID: PMC5948698.
Article14. Huggett JF, Cowen S, Foy CA. 2015; Considerations for digital PCR as an accurate molecular diagnostic tool. Clin Chem. 61:79–88. DOI: 10.1373/clinchem.2014.221366. PMID: 25338683.
Article15. Lion T, Watzinger F, Preuner S, Kreyenberg H, Tilanus M, de Weger R, et al. 2012; The EuroChimerism concept for a standardized approach to chimerism analysis after allogeneic stem cell transplantation. Leukemia. 26:1821–8. DOI: 10.1038/leu.2012.66. PMID: 22395360.
Article16. Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Ruf IK, Hindson BJ, et al. 2013; Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods. 10:1003–5. DOI: 10.1038/nmeth.2633. PMID: 23995387. PMCID: PMC4118677.
Article17. George D, Czech J, John B, Yu M, Jennings LJ. 2013; Detection and quantification of chimerism by droplet digital PCR. Chimerism. 4:102–8. DOI: 10.4161/chim.25400. PMID: 23974275. PMCID: PMC3782543.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Bone Marrow Chimerism Detection Using Next Generation Sequencing Based on Single Nucleotide Polymorphisms Following Liver Transplantation: Comparison With Short Tandem Repeat-PCR
- Mycobacterial infections in solid organ transplant recipients
- Management of Opportunistic Infections after Organ Transplantation
- The first case of brain death organ donation in a positive COVID-19 donors in Korea
- The Discrimination Power and Effectiveness of 3 Kinds of LTR Primers in the VNTR-PCR for Evaluation of the Engraftment of Allogeneic Peripheral Blood Stem Cells Transplantation