Ann Lab Med.
2021 Jul;41(4):394-400. 10.3343/alm.2021.41.4.394.
Accuracy and Performance Evaluation of Triplet Repeat Primed PCR as an Alternative to Conventional Diagnostic Methods for Fragile X Syndrome
- Affiliations
-
- 1Department of Laboratory Medicine, Graduate School, Kyung Hee University, Seoul, Korea
- 2Department of Laboratory Medicine, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea
- 3Department of Laboratory Medicine, Kosin University Gospel Hospital, Busan, Korea
- 4Biomedical Research Institute, Seoul National University Hospital, Seoul, Korea
- KMID: 2512719
- DOI: http://doi.org/10.3343/alm.2021.41.4.394
Abstract
- Background
Conventional diagnosis of fragile X syndrome (FXS) is based on a combination of fragment analysis (FA) and Southern blotting (SB); however, this diagnostic approach is time- and labor-intensive and has pitfalls such as the possibility of missing large number alleles. Triplet repeat primed PCR (TP-PCR) is a current alternative used to overcome these limitations. We evaluated the diagnostic usefulness of TP-PCR compared with the conventional diagnostic protocol consisting of FA and/or SB in terms of allele categorization, repeat number correlation, and zygosity concordance in female genetic carriers.
Methods
From November 2013 to March 2018, 458 patients (326 males, 132 females) were simultaneously examined using FA and/or SB and TP-PCR by detecting CGG repeat numbers in FMR gene and diagnosed as per American College of Medical Genetics guidelines.
Results
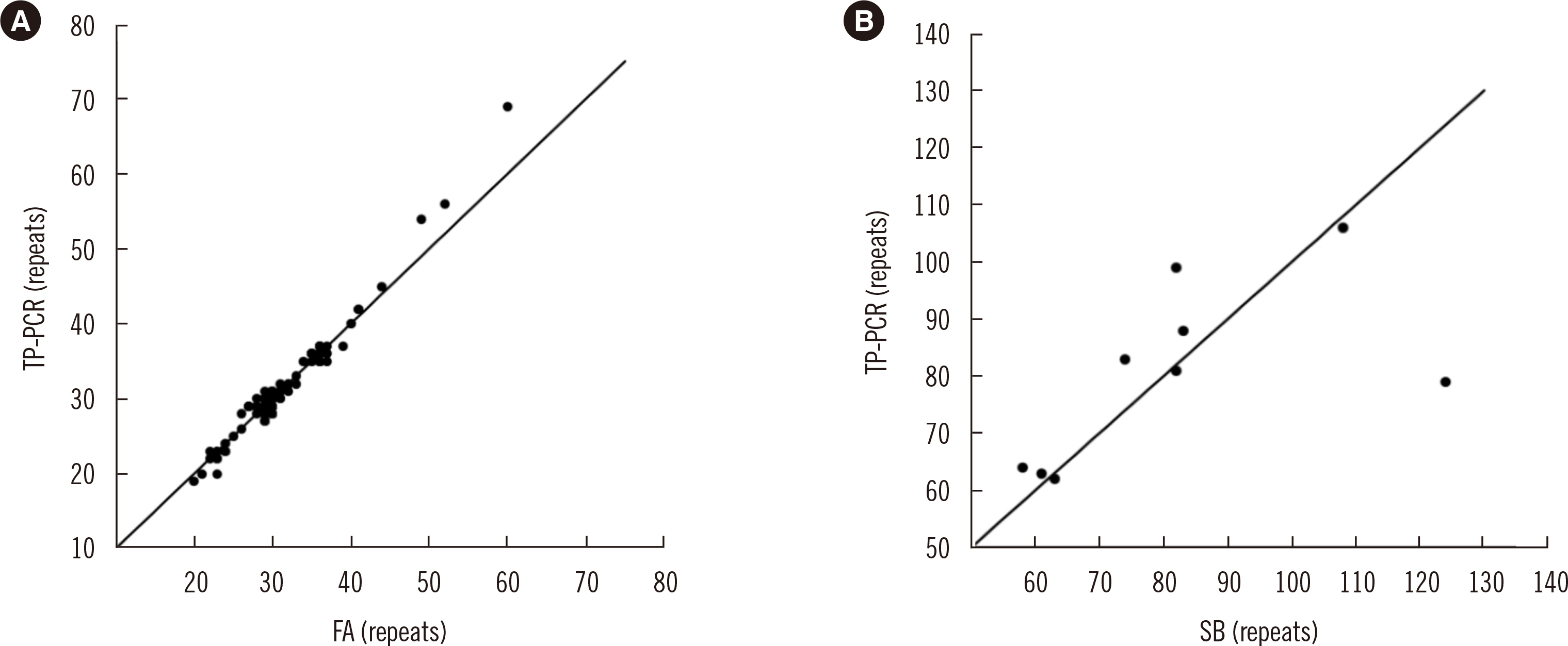
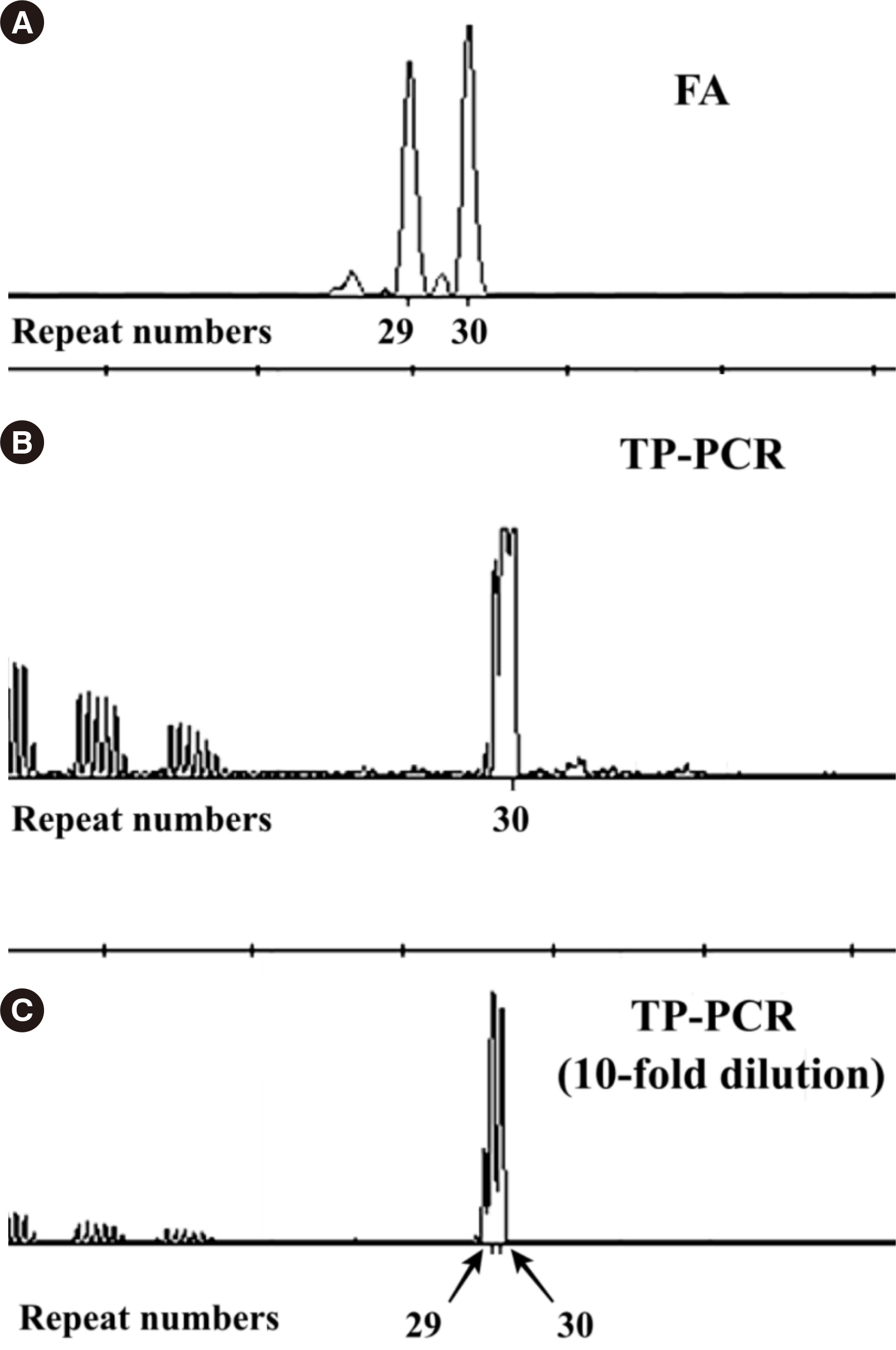
The TP-PCR results showed high concordance with the FA and/or SB results for all three aspects (allele categorization, repeat number correlation, and zygosity concordance in female genetic carriers). TP-PCR detected CGG expansions ≥ 200 in all full mutation (FM) allele cases in male patients, as well as both the normal allele (NL) and FM allele in female carriers. In premutation (PM) allele carriers, the TP-PCR results were consistent with the FA and/or SB results. In terms of zygosity concordance in female genetic carriers, 12 NL cases detected by TP-PCR showed a merged peak consisting of two close heterozygous peaks; however, this issue was resolved using a 10-fold dilution.
Conclusions
TP-PCR may serve as a reliable alternative method for FXS diagnosis.
Keyword
Figure
Reference
-
1. Hagerman RJ, Berry-Kravis E, Hazlett HC, Bailey DB Jr, Moine H, Kooy RF, et al. 2017; Fragile X syndrome. Nat Rev Dis Primers. 3:17065. DOI: 10.1038/nrdp.2017.65. PMID: 28960184.
Article2. Tassone , F . 2015; Advanced technologies for the molecular diagnosis of fragile X syndrome. Expert Rev Mol Diagn. 15:1465–73. DOI: 10.1586/14737159.2015.1101348. PMID: 26489042. PMCID: PMC4955806.
Article3. Monaghan KG, Lyon E, Spector EB. American College of Medical Genetics and Genomics. 2013; ACMG Standards and Guidelines for fragile X testing: a revision to the disease-specific supplements to the Standards and Guidelines for Clinical Genetics Laboratories of the. Genet Med. 15:575–86. DOI: 10.1038/gim.2013.61. PMID: 23765048.4. Biancalana V, Glaeser D, McQuaid S, Steinbach P. 2015; EMQN best practice guidelines for the molecular genetic testing and reporting of fragile X syndrome and other fragile X-associated disorders. Eur J Hum Genet. 23:417–25. DOI: 10.1038/ejhg.2014.185. PMID: 25227148. PMCID: PMC4666582.
Article5. Macpherson J, Sharif A. Practice guidelines for molecular diagnosis of fragile X syndrome. Association for Clinical Genetic Science (ACGS). https://www.acgs.uk.com/media/10768/frx_bpg_final_nov_2014.pdf. Updated on Nov 2014.6. Lyon E, Laver T, Yu P, Jama M, Young K, Zoccoli M, et al. 2010; A simple, high-throughput assay for Fragile X expanded alleles using triple repeat primed PCR and capillary electrophoresis. J Mol Diagn. 12:505–11. DOI: 10.2353/jmoldx.2010.090229. PMID: 20431035. PMCID: PMC2893636.
Article7. Tassone F, Pan R, Amiri K, Taylor AK, Hagerman PJ. 2008; A rapid polymerase chain reaction-based screening method for identification of all expanded alleles of the fragile X (FMR1) gene in newborn and high-risk populations. J Mol Diagn. 10:43–9. DOI: 10.2353/jmoldx.2008.070073. PMID: 18165273. PMCID: PMC2175542.8. Brykczynska U, Pecho-Vrieseling E, Thiemeyer A, Klein J, Fruh I, Doll T, et al. 2016; CGG repeat-induced FMR1 silencing depends on the expansion size in human iPSC's and neurons carrying unmethylated full mutations. Stem Cell Reports. 7:1059–71. DOI: 10.1016/j.stemcr.2016.10.004. PMID: 27840045. PMCID: PMC5161530.9. Jin Y, Lin B, Fung YS. 2001; Electrophoretic migration behavior of DNA fragments in polymer solution. Electrophoresis. 22:2150–8. DOI: 10.1002/1522-2683(20017)22:11<2150::AID-ELPS2150>3.0.CO;2-U.
Article10. Kiba Y, Zhang L, Baba Y. 2003; Anomalously fast migration of triplet‐repeat DNA in capillary electrophoresis with linear polymer solution. Electrophoresis. 24:452–7. DOI: 10.1002/elps.200390054. PMID: 12569536.
Article11. Jin P, Warren ST. 2000; Understanding the molecular basis of fragile X syndrome. Hum Mol Genet. 9:901–8. DOI: 10.1093/hmg/9.6.901. PMID: 10767313.
Article12. Nolin SL, Sah S, Glicksman A, Sherman SL, Allen E, Berry-Kravis E, et al. 2013; Fragile X AGG analysis provides new risk predictions for 45-69 repeat alleles. Am J Med Genet A. 161A:771–8. DOI: 10.1002/ajmg.a.35833. PMID: 23444167. PMCID: PMC4396070.
Article13. Sullivan AK, Marcus M, Epstein MP, Allen EG, Anido AE, Paquin JJ, et al. 2005; Association of FMR1 repeat size with ovarian dysfunction. Hum Reprod. 20:402–12. DOI: 10.1093/humrep/deh635. PMID: 15608041.14. Wakeling EN, Nahhas FA, Feldman GL. 2014; Extra alleles in FMR1 triple-primed PCR: artifact, aneuploidy, or somatic mosaicism? J Mol Diagn. 16:689–96. DOI: 10.1016/j.jmoldx.2014.06.006. PMID: 25307758.15. Godler DE, Tassone F, Loesch DZ, Taylor AK, Gehling F, Hagerman RJ, et al. 2010; Methylation of novel markers of fragile X alleles is inversely correlated with FMRP expression and FMR1 activation ratio. Hum Mol Genet. 19:1618–32. DOI: 10.1093/hmg/ddq037. PMID: 20118148. PMCID: PMC2846165.16. de Vries BB, Wiegers AM, Smits AP, Mohkamsing S, Duivenvoorden HJ, Fryns JP, et al. 1996; Mental status of females with an FMR1 gene full mutation. Am J Hum Genet. 58:1025–32.17. Loesch DZ, Sherwell S, Kinsella G, Tassone F, Taylor A, Amor D, et al. 2012; Fragile X-associated tremor/ataxia phenotype in a male carrier of unmethylated full mutation in the FMR1 gene. Clin Genet. 82:88–92. DOI: 10.1111/j.1399-0004.2011.01675.x. PMID: 21476992.18. Wöhrle D, Salat U, Gläser D, Mücke J, Meisel-Stosiek M, Schindler D, et al. 1998; Unusual mutations in high functioning fragile X males: apparent instability of expanded unmethylated CGG repeats. J Med Genet. 35:103–11. DOI: 10.1136/jmg.35.2.103. PMID: 9507388. PMCID: PMC1051212.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Molecular diagnosis of fragile X syndrome in a female child
- Molecular screening for fragile x syndrome in mentally handicapped children in korea
- Prenatal Population Screening for Fragile X Carrier and the Prevalence of Premutation Carriers in Korea
- Incidence of Fragile X Syndrome in Korean Patients with Mental Retardation
- A Study on Prevalence of Premutation Sized FMR1 Gene Using Polymerase Chain Reaction