J Korean Med Sci.
2021 Jan;36(4):e38. 10.3346/jkms.2021.36.e38.
Epidemiologic Linkage of COVID-19 Outbreaks at Two University-affiliated Hospitals in the Seoul Metropolitan Area in March 2020
- Affiliations
-
- 1Department of Laboratory Medicine, University of Ulsan College of Medicine and Asan Medical Center, Seoul, Korea
- 2Department of Laboratory Medicine, Uijeongbu St. Mary's Hospital, College of Medicine, The Catholic University of Korea, Uijeongbu, Korea
- 3ChunLab Inc., Seoul, Korea
- 4Department of infectious disease, University of Ulsan College of Medicine and Asan Medical Center, Seoul, Korea
- 5Department of Pediatrics, University of Ulsan College of Medicine and Asan Medical Center, Seoul, Korea
- 6Department of Biological Sciences, Seoul National University, Seoul, Korea
- KMID: 2511636
- DOI: http://doi.org/10.3346/jkms.2021.36.e38
Abstract
- Background
Coronavirus disease 2019 (COVID-19) outbreaks emerged at two universityaffiliated hospitals in Seoul (hospital A) and Uijeongbu City (hospital S) in the metropolitan Seoul area in March 2020. The aim of this study was to investigate epidemiological links between the outbreaks using whole genome sequencing (WGS) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
Methods
Fifteen patients were enrolled in the study, including four non-outbreak (A1–A4) and three outbreak cases (A5–A7) in hospital A and eight cases (S1–S8) in hospital S. Patients' hospital stays, COVID-19 symptoms, and transfer history were reviewed. RNA samples were submitted for WGS and genome-wide single nucleotide variants and phylogenetic relationships were analyzed.
Results
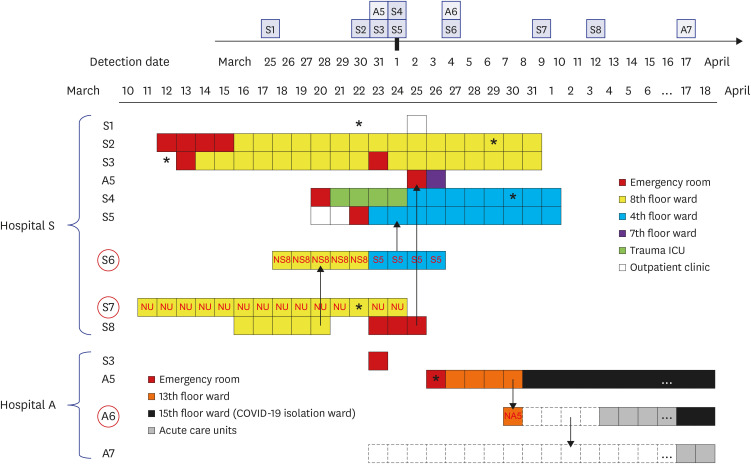
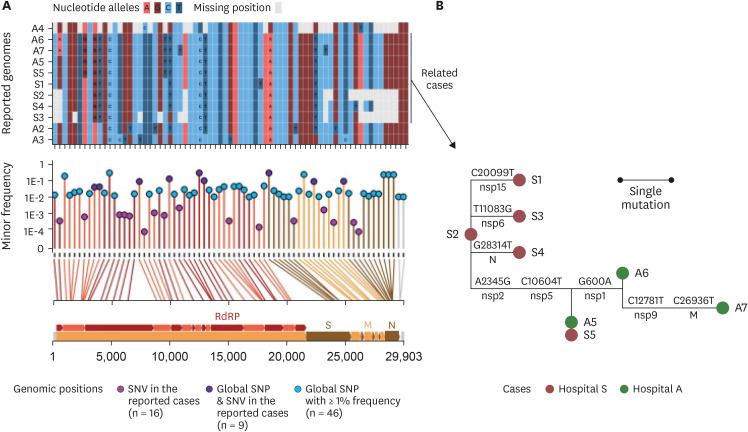
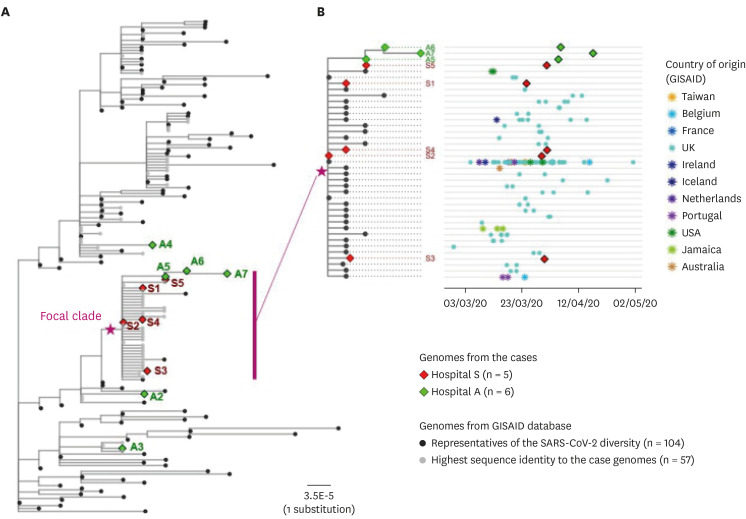
The index patient (A5) in hospital A was transferred from hospital S on 26 March. Patients A6 and A7 were the family caregiver and sister, respectively, of the patient who shared a room with A5 for 4 days. Prior to transfer, A5 was at the next bed to S8 in the emergency room on 25 March. Patient S6, a professional caregiver, took care of the patient in the room next to S8's room for 5 days until 22 March and then S5 for another 3 days. WGS revealed that SARS-CoV-2 in A2, A3, and A4 belong to clades V/B.2, S/A, and G/B.1, respectively, whereas that of A5–A7 and S1-S5 are of the V/B.2.1 clade and closely clustered. In particular, SARS-CoV-2 in patients A5 and S5 showed perfect identity.
Conclusion
WGS is a useful tool to understand epidemiology of SARS-CoV-2. It is the first study to elucidate the role of patient transfer and caregivers as links of nosocomial outbreaks of COVID-19 in multiple hospitals.
Keyword
Figure
Reference
-
1. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020; 395(10223):497–506. PMID: 31986264.
Article2. Weekly operational update on COVID-19 - 21 December 2020. Updated 2020. Accessed December 22, 2020. https://www.who.int/publications/m/item/weekly-operational-update-on-covid-19---21-december-2020.3. Kim JY, Choe PG, Oh Y, Oh KJ, Kim J, Park SJ, et al. The first case of 2019 novel coronavirus pneumonia imported into Korea from Wuhan, China: implication for infection prevention and control measures. J Korean Med Sci. 2020; 35(5):e61. PMID: 32030925.
Article4. The updates on COVID-19 in Korea as of 21 December. Updated 2020. Accessed December 22, 2020. https://www.cdc.go.kr/board/board.es?mid=a30402000000&bid=0030.5. The updates on COVID-19 in Korea as of 6 July. Updated 2020. Accessed July 6, 2020. http://www.cdc.kr/board/board.es?mid=a30402000000&bid=0030.6. COVID-19 National Emergency Response Center. Epidemiology and Case Management Team. Korea Centers for Disease Control and Prevention. Coronavirus disease-19: the first 7,755 cases in the Republic of Korea. Osong Public Health Res Perspect. 2020; 11(2):85–90. PMID: 32257774.7. The updates on COVID-19 in Korea as of 8 July. Updated 2020. Accessed July 8, 2020. https://www.cdc.go.kr/board/board.es?mid=a30402000000&bid=0030.8. Park SY, Kim YM, Yi S, Lee S, Na BJ, Kim CB, et al. Coronavirus disease outbreak in call center, South Korea. Emerg Infect Dis. 2020; 26(8):1666–1670. PMID: 32324530.
Article9. Guo W, Li M, Dong Y, Zhou H, Zhang Z, Tian C, et al. Diabetes is a risk factor for the progression and prognosis of COVID-19. Diabetes Metab Res Rev. 2020; 36(7):e3319.
Article10. Wang X, Zhou Q, He Y, Liu L, Ma X, Wei X, et al. Nosocomial outbreak of COVID-19 pneumonia in Wuhan, China. Eur Respir J. 2020; 55(6):2000544. PMID: 32366488.
Article11. Schwierzeck V, König JC, Kühn J, Mellmann A, Correa-Martínez CL, Omran H, et al. First reported nosocomial outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in a pediatric dialysis unit. Clin Infect Dis. 2020; ciaa491. PMID: 32337584.12. Dudas G, Bedford T. The ability of single genes vs full genomes to resolve time and space in outbreak analysis. BMC Evol Biol. 2019; 19(1):232. PMID: 31878875.
Article13. Grubaugh ND, Faria NR, Andersen KG, Pybus OG. genomic insights into zika virus emergence and spread. Cell. 2018; 172(6):1160–1162. PMID: 29522736.
Article14. Clade and lineage nomenclature, July 4, 2020. Updated 2020. Accessed July 7, 2020. https://www.gisaid.org/references/statements-clarifications/clade-and-lineage-nomenclature-aids-in-genomic-epidemiology-of-active-hcov-19-viruses/.15. Rambaut A, Holmes EC, O'Toole Á, Hill V, McCrone JT, Ruis C, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020; 5(11):1403–1407. PMID: 32669681.
Article16. Böhmer MM, Buchholz U, Corman VM, Hoch M, Katz K, Marosevic DV, et al. Investigation of a COVID-19 outbreak in Germany resulting from a single travel-associated primary case: a case series. Lancet Infect Dis. 2020; 20(8):920–928. PMID: 32422201.
Article17. Sekizuka T, Itokawa K, Kageyama T, Saito S, Takayama I, Asanuma H, et al. Haplotype networks of SARS-CoV-2 infections in the Diamond Princess cruise ship outbreak. Proc Natl Acad Sci U S A. 2020; 117(33):20198–20201. PMID: 32723824.
Article18. Meredith LW, Hamilton WL, Warne B, Houldcroft CJ, Hosmillo M, Jahun AS, et al. Rapid implementation of SARS-CoV-2 sequencing to investigate cases of health-care associated COVID-19: a prospective genomic surveillance study. Lancet Infect Dis. 2020; 20(11):1263–1272. PMID: 32679081.
Article19. Jung J, Hong MJ, Kim EO, Lee J, Kim MN, Kim SH. Investigation of a nosocomial outbreak of coronavirus disease 2019 in a paediatric ward in South Korea: successful control by early detection and extensive contact tracing with testing. Clin Microbiol Infect. 2020; 26(11):1574–1575. PMID: 32593744.
Article20. Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020; 579(7798):265–269. PMID: 32015508.
Article21. Shu Y, McCauley J. GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017; 22(13):30494. PMID: 28382917.
Article22. Rhee C, Baker M, Vaidya V, Tucker R, Resnick A, Morris CA, et al. Incidence of nosocomial COVID-19 in patients hospitalized at a large US academic medical center. JAMA Netw Open. 2020; 3(9):e2020498. PMID: 32902653.
Article23. Ran L, Chen X, Wang Y, Wu W, Zhang L, Tan X. Risk factors of healthcare workers with corona virus disease 2019: a retrospective cohort study in a designated hospital of Wuhan in China. Clin Infect Dis. 2020; 71(16):2218–2221. PMID: 32179890.24. Dong Y, Mo X, Hu Y, Qi X, Jiang F, Jiang Z, et al. Epidemiology of COVID-19 among children in China. Pediatrics. 2020; 145(6):e20200702. PMID: 32179660.
Article25. Qiu H, Wu J, Hong L, Luo Y, Song Q, Chen D. Clinical and epidemiological features of 36 children with coronavirus disease 2019 (COVID-19) in Zhejiang, China: an observational cohort study. Lancet Infect Dis. 2020; 20(6):689–696. PMID: 32220650.
Article26. Bai Y, Yao L, Wei T, Tian F, Jin DY, Chen L, et al. Presumed asymptomatic carrier transmission of COVID-19. JAMA. 2020; 323(14):1406–1407. PMID: 32083643.
Article27. Yamagishi T, Kamiya H, Kakimoto K, Suzuki M, Wakita T. Descriptive study of COVID-19 outbreak among passengers and crew on Diamond Princess cruise ship, Yokohama Port, Japan, 20 January to 9 February 2020. Euro Surveill. 2020; 25(23):28–35.
Article28. Xu XW, Wu XX, Jiang XG, Xu KJ, Ying LJ, Ma CL, et al. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: retrospective case series. BMJ. 2020; 368:m606. PMID: 32075786.
Article29. Sutton D, Fuchs K, D'Alton M, Goffman D. Universal screening for SARS-CoV-2 in women admitted for delivery. N Engl J Med. 2020; 382(22):2163–2164. PMID: 32283004.
Article30. Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, et al. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020; 182(4):812–827.e19. PMID: 32697968.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Coronavirus Disease 2019 Cases at Universities and Colleges in Seoul Metropolitan Area
- Report on the Epidemiological Features of Coronavirus Disease 2019 (COVID-19) Outbreak in the Republic of Korea from January 19 to March 2, 2020
- Experience of Treating Critically Ill COVID-19 Patients in Daegu, South Korea
- Coronavirus Disease-19: The First 7,755 Casesin the Republic of Korea
- Analysis on 54 Mortality Cases of Coronavirus Disease 2019 in the Republic of Korea from January 19 to March 10, 2020