Cancer Res Treat.
2021 Jan;53(1):148-161. 10.4143/crt.2020.424.
An Innovative Prognostic Model Based on Four Genes in Asian Patient with Gastric Cancer
- Affiliations
-
- 1Department of Gastrointestinal Surgery, Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education), Peking University Cancer Hospital and Institute, Beijing, China
- 2Department of Gastrointestinal Surgery, Shandong Provincial Hospital, Cheeloo College of Medicine, Shandong University, Jinan, China
- 3First Affiliated Hospital of Baotou Medical College, General Surgery, Baotou, China
- 4Department of Gastrointestinal Surgery, Peking University Shenzhen Hospital, Shenzhen, China
- KMID: 2510657
- DOI: http://doi.org/10.4143/crt.2020.424
Abstract
- Purpose
Gastric cancer (GC) has substantial biological differences between Asian and non-Asian populations, which makes it difficult to have a unified predictive measure for all people. We aimed to identify novel prognostic biomarkers to help predict the prognosis of Asian GC patients.
Materials and Methods
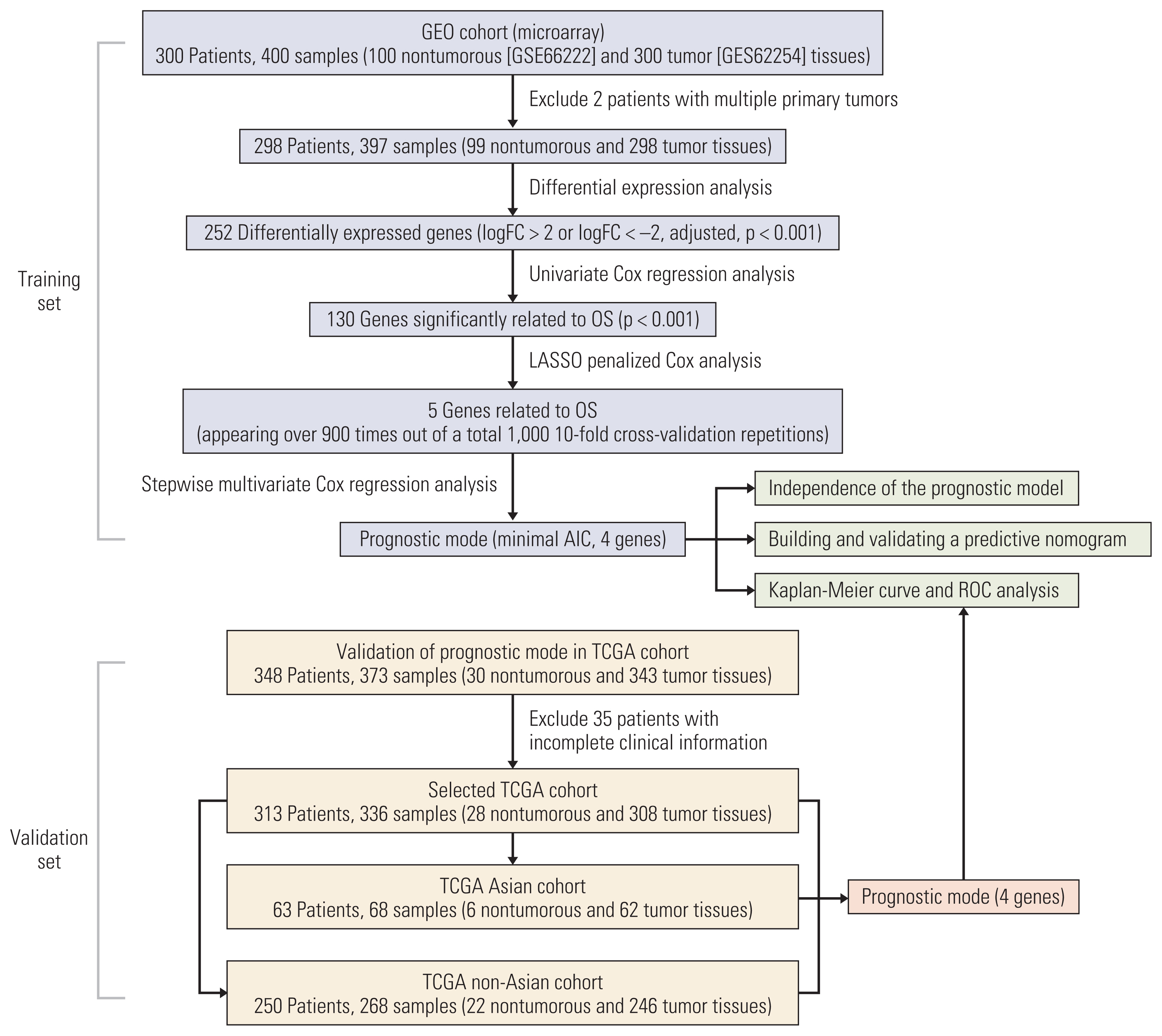
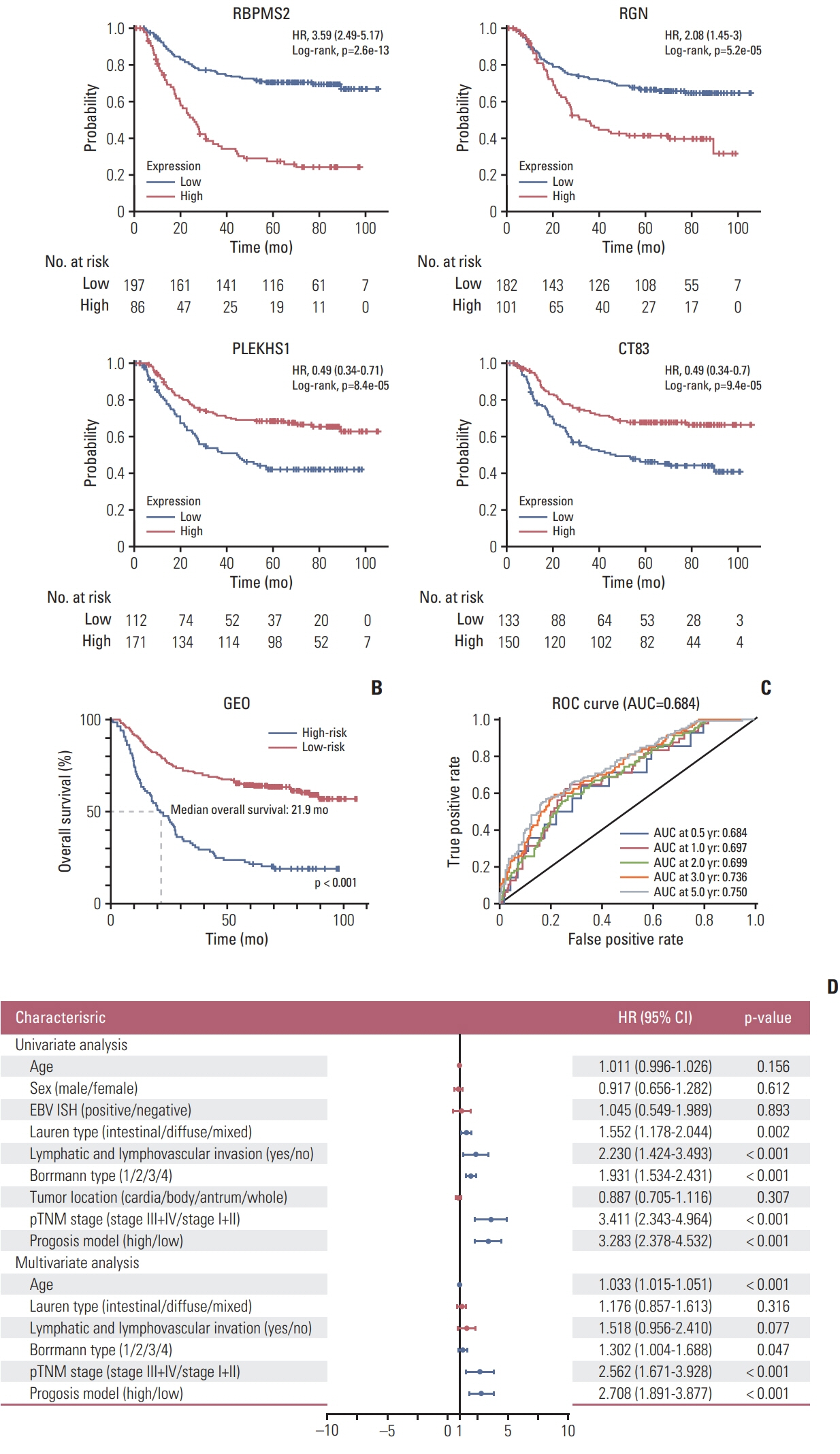
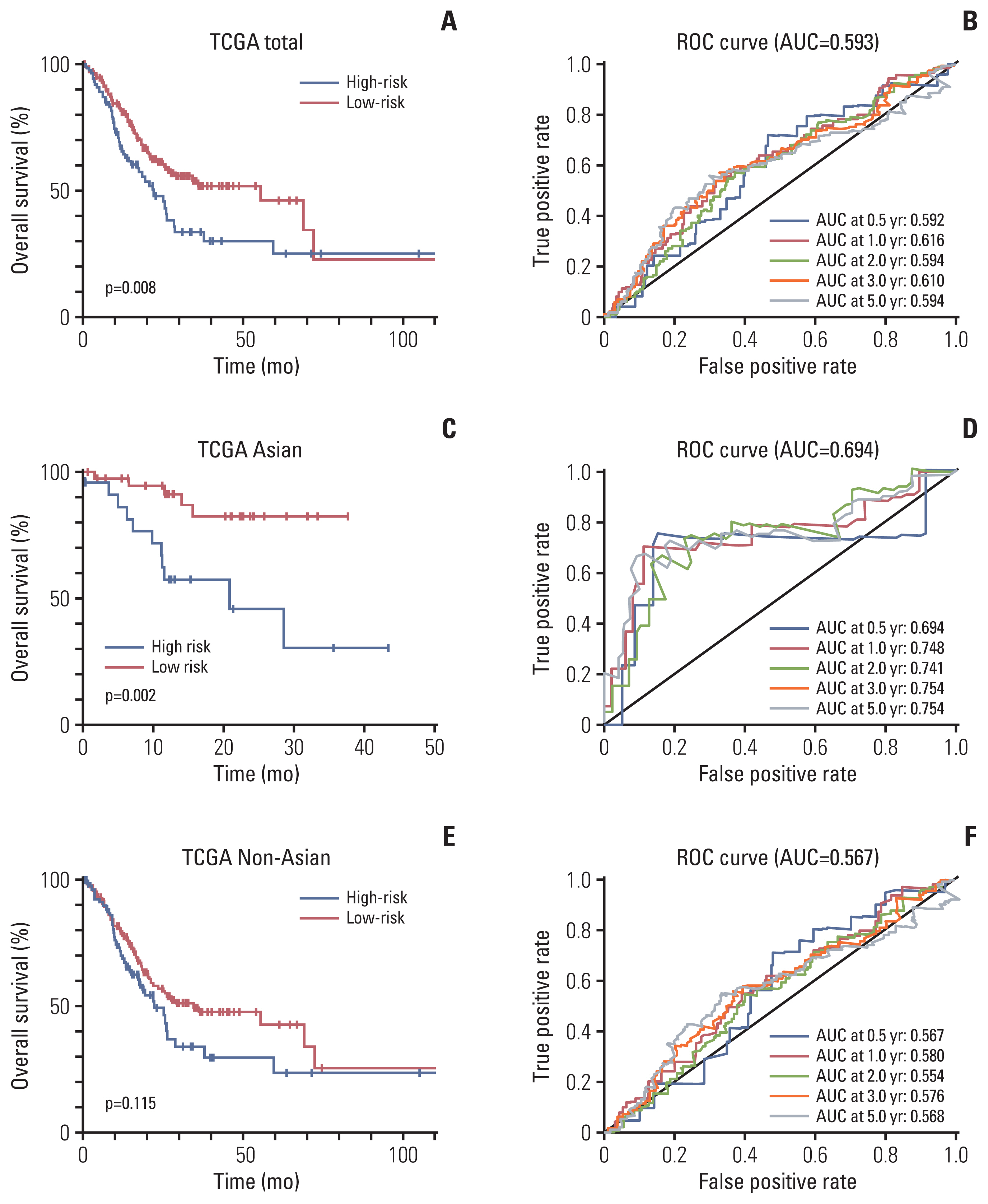
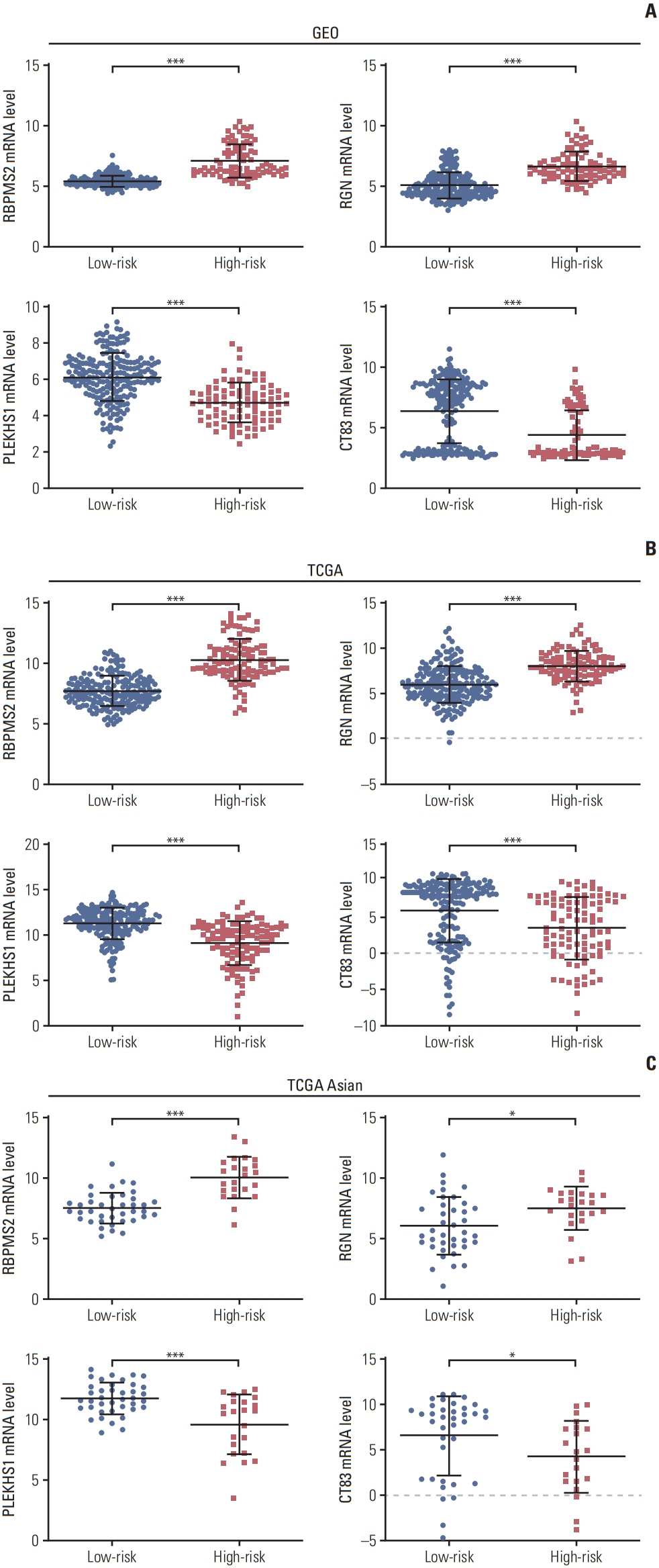
We investigated the differential gene expression between GC and normal tissues of GSE66229. Univariate, multivariate and Lasso Cox regression analyses were conducted to establish a four-gene-related prognostic model based on the risk score. The risk score was based on a linear combination of the expression levels of individual genes multiplied by their multivariate Cox regression coefficients. Validation of the prognostic model was conducted using The Cancer Genome Atlas (TCGA) database. A nomogram containing clinical characteristics and the prognostic model was established to predict the prognosis of Asian GC patients.
Results
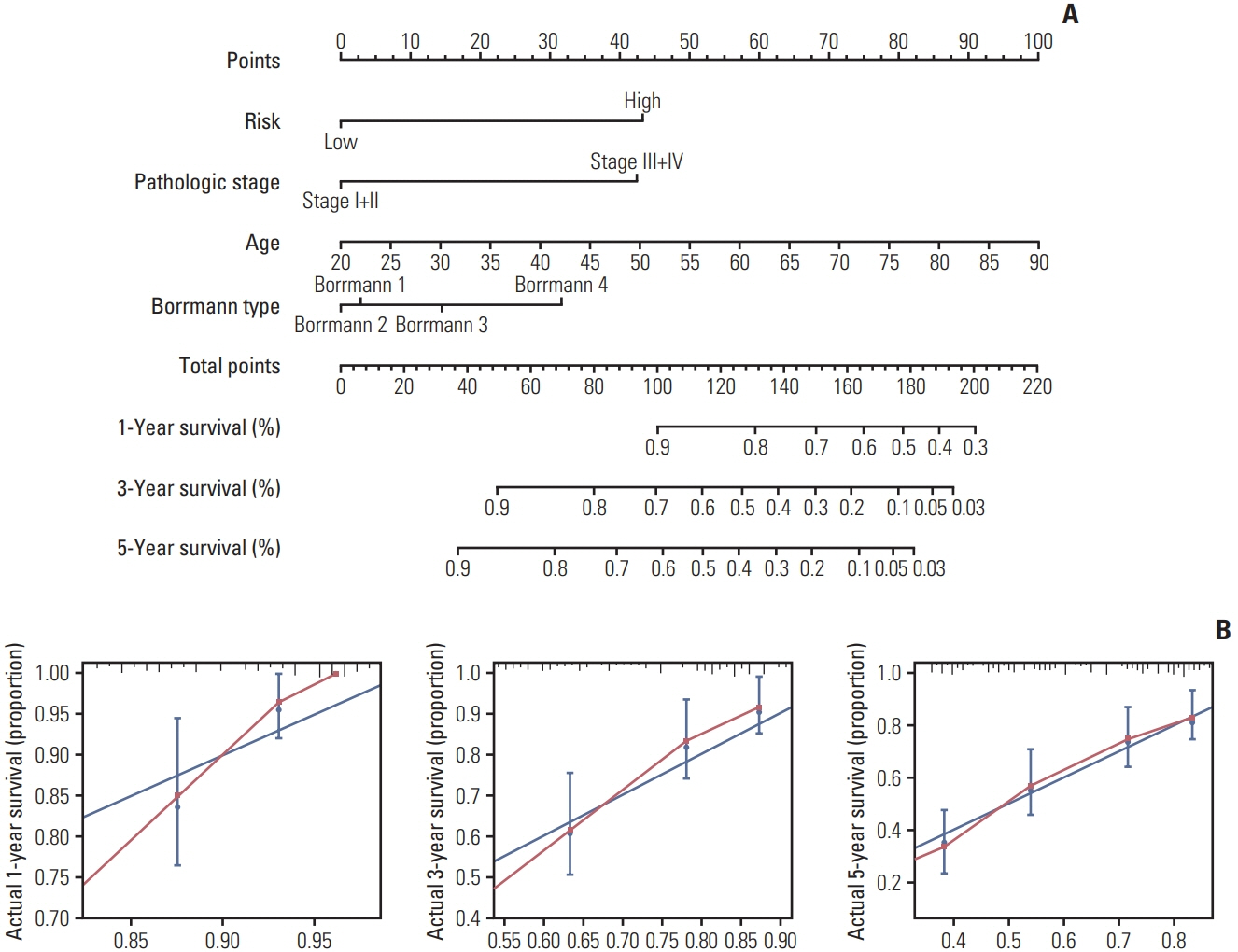
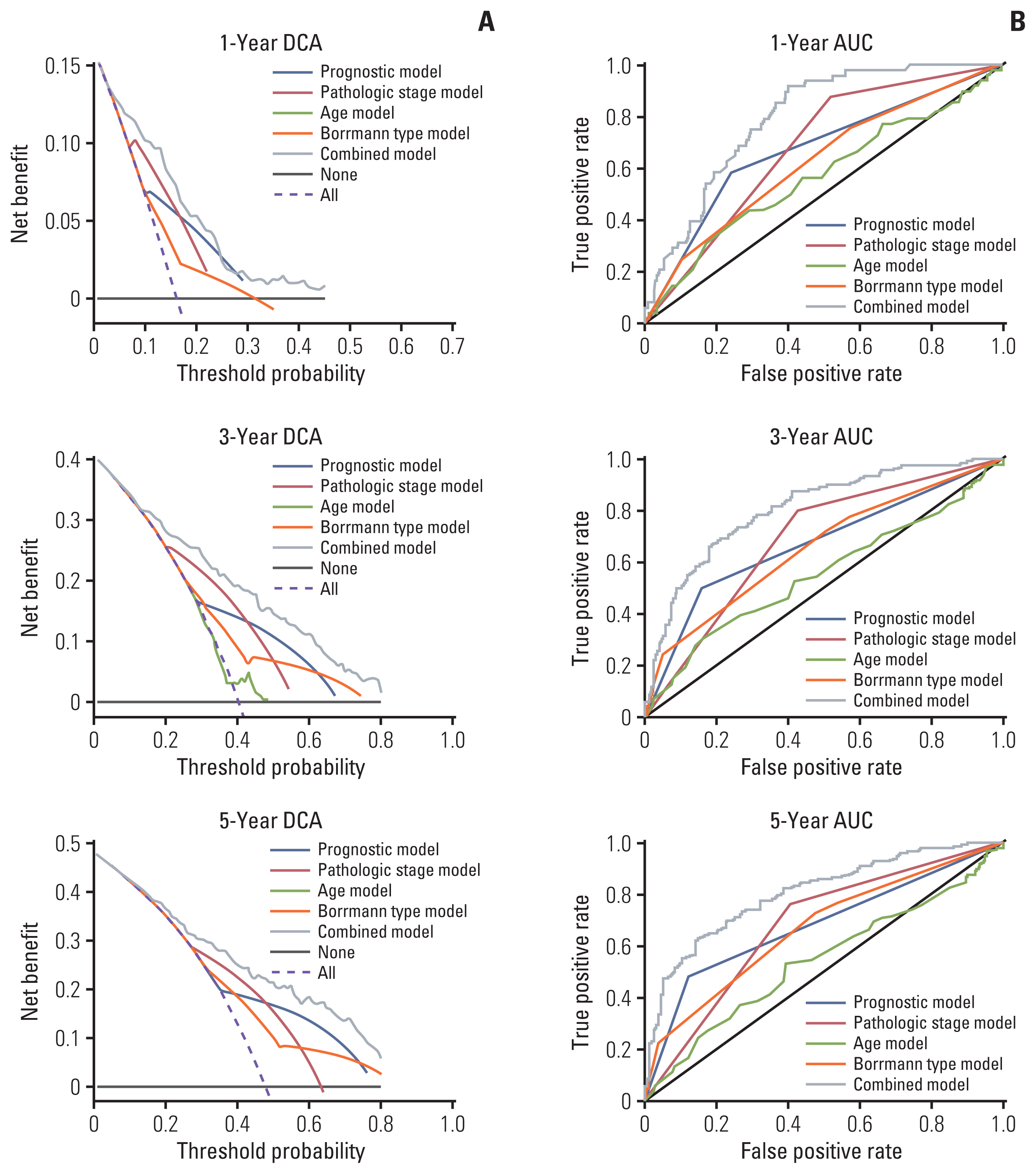
Four genes (RBPMS2, RGN, PLEKHS1, and CT83) were selected to establish the prognostic model, and it was validated in the TCGA Asian cohort. Receiver operating characteristic analysis confirmed the sensitivity and specificity of the prognostic model. Based on the prognostic model, a nomogram containing clinical characteristics and the prognostic model was established, and Harrell’s concordance index of the nomogram for evaluating the overall survival significantly higher than the model only focuses on the pathologic stage (0.74 vs. 0.64, p < 0.001).
Conclusion
The four-gene-related prognostic model and the nomogram based on it are reliable tools for predicting the overall survival of Asian GC patients.
Figure
Reference
-
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018; 68:394–424.
Article2. National Health Commission of The People’s Republic of China. Chinese guidelines for diagnosis and treatment of gastric cancer 2018 (English version). Chin J Cancer Res. 2019; 31:707–37.3. Chen X, Liu H, Li G, Yu J. Implications of clinical research on adjuvant chemotherapy for gastric cancer: where to go next? Chin J Cancer Res. 2019; 31:892–900.
Article4. Lin SJ, Gagnon-Bartsch JA, Tan IB, Earle S, Ruff L, Pettinger K, et al. Signatures of tumour immunity distinguish Asian and non-Asian gastric adenocarcinomas. Gut. 2015; 64:1721–31.
Article5. Long J, Zhang L, Wan X, Lin J, Bai Y, Xu W, et al. A four-gene-based prognostic model predicts overall survival in patients with hepatocellular carcinoma. J Cell Mol Med. 2018; 22:5928–38.
Article6. Hou JY, Wang YG, Ma SJ, Yang BY, Li QP. Identification of a prognostic 5-Gene expression signature for gastric cancer. J Cancer Res Clin Oncol. 2017; 143:619–29.
Article7. Cheong JH, Yang HK, Kim H, Kim WH, Kim YW, Kook MC, et al. Predictive test for chemotherapy response in resectable gastric cancer: a multi-cohort, retrospective analysis. Lancet Oncol. 2018; 19:629–38.
Article8. Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015; 21:449–56.
Article9. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47.
Article10. __Long J, Wang A, Bai Y, Lin J, Yang X, Wang D, et al. Development and validation of a TP53-associated immune prognostic model for hepatocellular carcinoma. EBioMedicine. 2019; 42:363–74.
Article11. Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004; 10:7252–9.12. Balachandran VP, Gonen M, Smith JJ, DeMatteo RP. Nomograms in oncology: more than meets the eye. Lancet Oncol. 2015; 16:e173–80.
Article13. Iasonos A, Schrag D, Raj GV, Panageas KS. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol. 2008; 26:1364–70.
Article14. Chen S, Chen X, Nie R, Ou Yang L, Liu A, Li Y, et al. A nomogram to predict prognosis for gastric cancer with peritoneal dissemination. Chin J Cancer Res. 2018; 30:449–59.
Article15. Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making. 2006; 26:565–74.
Article16. Sano T, Coit DG, Kim HH, Roviello F, Kassab P, Wittekind C, et al. Proposal of a new stage grouping of gastric cancer for TNM classification: International Gastric Cancer Association staging project. Gastric Cancer. 2017; 20:217–25.
Article17. Zhang J, Yuan Y, Wei Z, Ren J, Hou X, Yang D, et al. Crosstalk between prognostic long noncoding RNAs and messenger RNAs as transcriptional hallmarks in gastric cancer. Epigenomics. 2018; 10:433–43.
Article18. Jin H, Pinheiro PS, Callahan KE, Altekruse SF. Examining the gastric cancer survival gap between Asians and whites in the United States. Gastric Cancer. 2017; 20:573–82.
Article19. Kolonel LN, Hankin JH, Nomura AM. Multiethnic studies of diet, nutrition, and cancer in Hawaii. Princess Takamatsu Symp. 1985; 16:29–40.20. Sagnol S, Yang Y, Bessin Y, Allemand F, Hapkova I, Notarnicola C, et al. Homodimerization of RBPMS2 through a new RRM-interaction motif is necessary to control smooth muscle plasticity. Nucleic Acids Res. 2014; 42:10173–84.
Article21. Notarnicola C, Rouleau C, Le Guen L, Virsolvy A, Richard S, Faure S, et al. The RNA-binding protein RBPMS2 regulates development of gastrointestinal smooth muscle. Gastroenterology. 2012; 143:687–97.
Article22. Hapkova I, Skarda J, Rouleau C, Thys A, Notarnicola C, Janikova M, et al. High expression of the RNA-binding protein RBPMS2 in gastrointestinal stromal tumors. Exp Mol Pathol. 2013; 94:314–21.
Article23. Yamaguchi M, Osuka S, Shoji M, Weitzmann MN, Murata T. Survival of lung cancer patients is prolonged with higher regucalcin gene expression: suppressed proliferation of lung adenocarcinoma A549 cells in vitro. Mol Cell Biochem. 2017; 430:37–46.
Article24. Yamaguchi M, Osuka S, Weitzmann MN, Shoji M, Murata T. Increased regucalcin gene expression extends survival in breast cancer patients: overexpression of regucalcin suppresses the proliferation and metastatic bone activity in MDA-MB-231 human breast cancer cells in vitro. Int J Oncol. 2016; 49:812–22.
Article25. Weinhold N, Jacobsen A, Schultz N, Sander C, Lee W. Genome-wide analysis of noncoding regulatory mutations in cancer. Nat Genet. 2014; 46:1160–5.
Article26. Liu X, Wu J, Zhang D, Bing Z, Tian J, Ni M, et al. Identification of potential key genes associated with the pathogenesis and prognosis of gastric cancer based on integrated bioinformatics analysis. Front Genet. 2018; 9:265.
Article27. Marcinkowski B, Stevanovic S, Helman SR, Norberg SM, Serna C, Jin B, et al. Cancer targeting by TCR gene-engineered T cells directed against Kita-Kyushu Lung Cancer Antigen-1. J Immunother Cancer. 2019; 7:229.
Article28. Paret C, Simon P, Vormbrock K, Bender C, Kolsch A, Breitkreuz A, et al. CXorf61 is a target for T cell based immunotherapy of triple-negative breast cancer. Oncotarget. 2015; 6:25356–67.
Article29. Shigematsu Y, Hanagiri T, Shiota H, Kuroda K, Baba T, Mizukami M, et al. Clinical significance of cancer/testis antigens expression in patients with non-small cell lung cancer. Lung Cancer. 2010; 68:105–10.
Article30. Futawatari N, Fukuyama T, Yamamura R, Shida A, Takahashi Y, Nishi Y, et al. Early gastric cancer frequently has high expression of KK-LC-1, a cancer-testis antigen. World J Gastroenterol. 2017; 23:8200–6.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Clinicopathological Features and Survival of Patients with Gastric Cancer with a Family History: a Large Analysis of 2,736 Patients with Gastric Cancer
- Neoadjuvant Chemotherapy in Asian Patients With Locally Advanced Gastric Cancer
- The Clinical Significance of Mutation in the p53, DCC and nm23 Genes in Patients with Gastric Carcinoma
- Tumor Size as a Prognostic Factor in Gastric Cancer Patient
- Lymph Node Ratio System for N Staging of Gastric Cancer: Challenging for Universal Application But Useful for the Prognostic Prediction of Individual Patients