Clin Exp Otorhinolaryngol.
2020 Nov;13(4):326-339. 10.21053/ceo.2020.00654.
Recent Advances in the Application of Artificial Intelligence in Otorhinolaryngology-Head and Neck Surgery
- Affiliations
-
- 1Department of Mechanical Engineering, Pohang University of Science and Technology, Pohang, Korea
- 2Department of Otolaryngology-Head and Neck Surgery, Seoul St. Mary’s Hospital, College of Medicine, The Catholic University of Korea, Seoul, Korea
- 3Graduate School of Artificial Intelligence, Pohang University of Science and Technology, Pohang, Korea
- KMID: 2508510
- DOI: http://doi.org/10.21053/ceo.2020.00654
Abstract
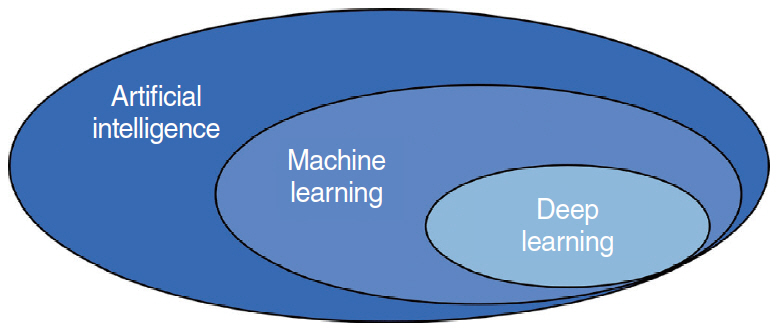
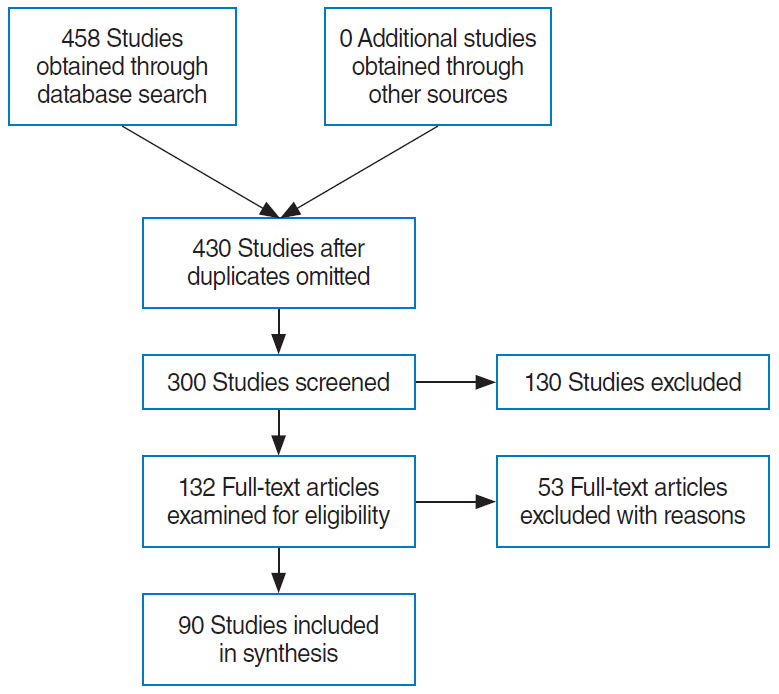
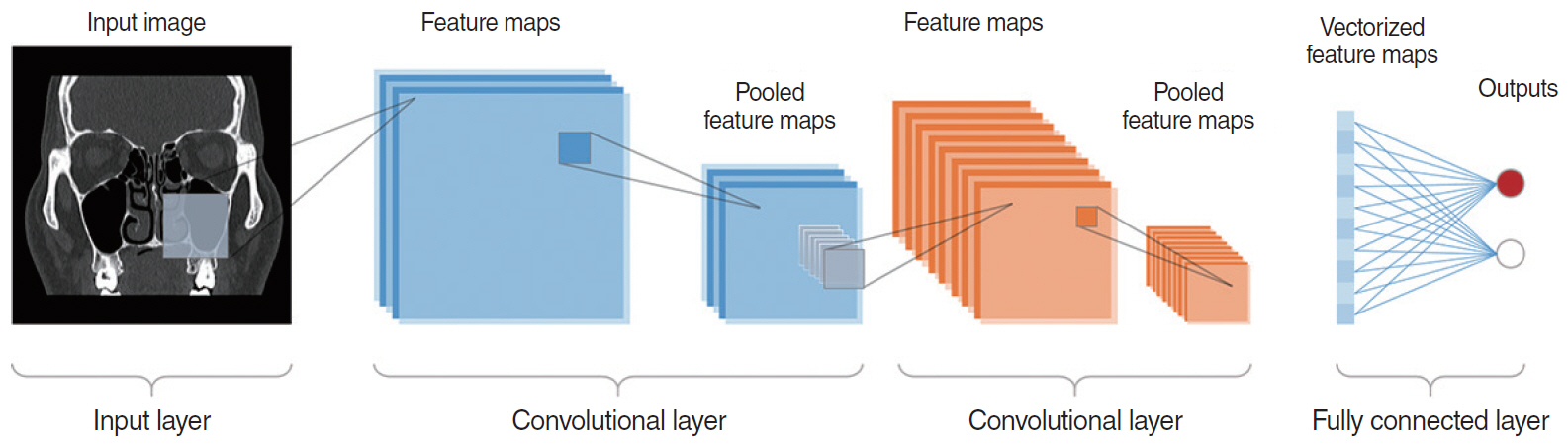
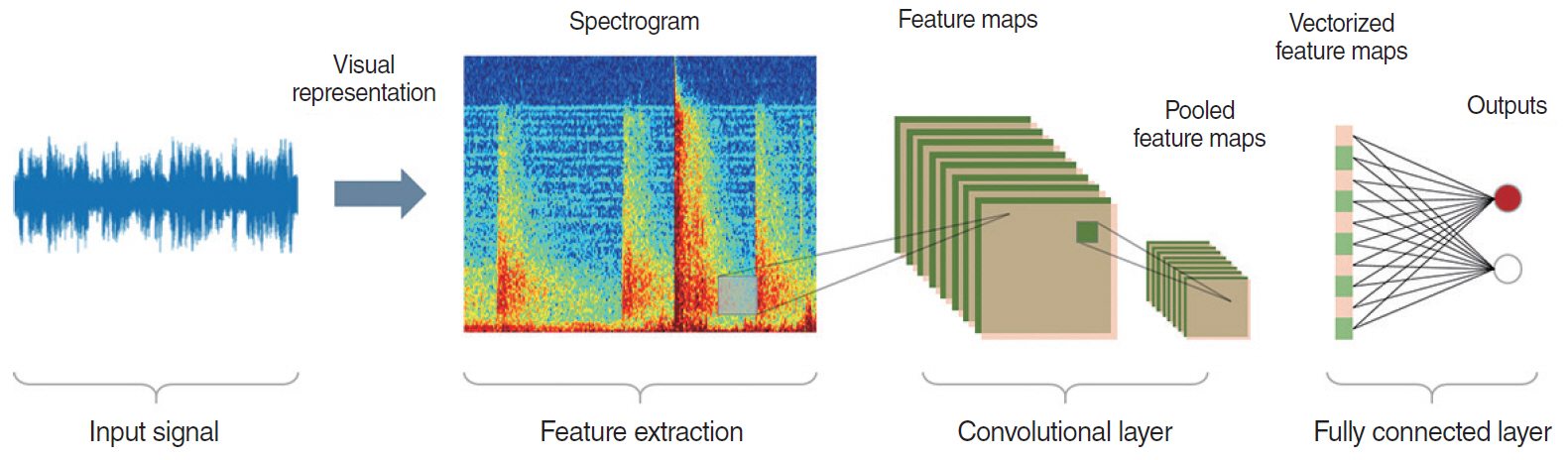
- This study presents an up-to-date survey of the use of artificial intelligence (AI) in the field of otorhinolaryngology, considering opportunities, research challenges, and research directions. We searched PubMed, the Cochrane Central Register of Controlled Trials, Embase, and the Web of Science. We initially retrieved 458 articles. The exclusion of non-English publications and duplicates yielded a total of 90 remaining studies. These 90 studies were divided into those analyzing medical images, voice, medical devices, and clinical diagnoses and treatments. Most studies (42.2%, 38/90) used AI for image-based analysis, followed by clinical diagnoses and treatments (24 studies). Each of the remaining two subcategories included 14 studies. Machine learning and deep learning have been extensively applied in the field of otorhinolaryngology. However, the performance of AI models varies and research challenges remain.
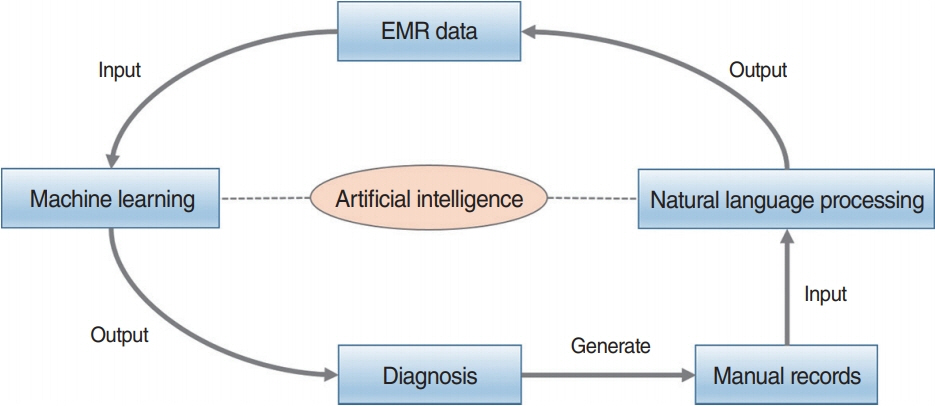
Figure
Reference
-
1. Pomerol JC. Artificial intelligence and human decision making. Eur J Oper Res. 1997; May. 99(1):3–25.
Article2. Simon HA. The sciences of the artificial. Cambridge (MA): MIT Press;2019.3. In : He K, Zhang X, Ren S, Sun J, editors. Deep residual learning for image recognition. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2016.
Article4. Silver D, Hassabis D, editors. Alphago: mastering the ancient game of go with machine learning [Internet]. Google AI Blog;2016. [cited 2020 Aug 1]. Available from: https://ai.googleblog.com/2016/01/alphagomastering-ancient-game-of-go.html.5. Densen P. Challenges and opportunities facing medical education. Trans Am Clin Climatol Assoc. 2011; 122:48–58.6. Monegain B, editor. IBM Watson pinpoints rare form of leukemia after doctors misdiagnosed patient [Internet]. Healthcare IT News;2016. [cited 2020 Aug 1]. Available from: https://www.healthcareitnews.com/news/ibm-watson-pinpoints-rare-form-leukemia-after-doctors-misdiagnosed-patient.7. Grand View Research. Artificial intelligence in healthcare market size, share & trends analysis by component (hardware, software, services), by application, by region, competitive insights, and segment forecasts, 2019-2025 [Internet]. San Francisco, CA: Grand View Research;2019. [cited 2020 Aug 1]. Available from: https://www.grandviewresearch.com/industry-analysis/artificial-intelligence-ai-healthcare-market.8. Crowson MG, Ranisau J, Eskander A, Babier A, Xu B, Kahmke RR, et al. A contemporary review of machine learning in otolaryngology-head and neck surgery. Laryngoscope. 2020; Jan. 130(1):45–51.
Article9. Freeman DT, editor. Computer recognition of brain stem auditory evoked potential wave V by a neural network. Proc Annu Symp Comput Appl Med Care. 1991; 305–9.10. Zadak J, Unbehauen R. An application of mapping neural networks and a digital signal processor for cochlear neuroprostheses. Biol Cybern. 1993; 68(6):545–52.11. Kim DH, Kim Y, Park JS, Kim SW. Virtual reality simulators for endoscopic sinus and skull base surgery: the present and future. Clin Exp Otorhinolaryngol. 2019; Feb. 12(1):12–7.
Article12. Park JH, Jeon HJ, Lim EC, Koo JW, Lee HJ, Kim HJ, et al. Feasibility of eye tracking assisted vestibular rehabilitation strategy using immersive virtual reality. Clin Exp Otorhinolaryngol. 2019; Nov. 12(4):376–84.
Article13. Song JJ. Virtual reality for vestibular rehabilitation. Clin Exp Otorhinolaryngol. 2019; Nov. 12(4):329–30.
Article14. Chollet F, Allaire JJ. Deep learning with R. Shelter Island (NY): Manning Publications;2018.15. Mohri M, Rostamizadeh A, Talwalkar A. Foundations of machine learning. Cambridge (MA): MIT Press;2012.16. Flach P. Machine learning: the art and science of algorithms that make sense of data. Cambridge: Cambridge University Press;2012.17. James G, Witten D, Hastie T, Tibshirani R. An introduction to statistical learning. Berlin: Springer;2013.18. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015; May. 521(7553):436–44.
Article19. Tama BA, Rhee KH. Tree-based classifier ensembles for early detection method of diabetes: an exploratory study. Artif Intell Rev. 2019; 51:355–70.
Article20. Huang J, Habib AR, Mendis D, Chong J, Smith M, Duvnjak M, et al. An artificial intelligence algorithm that differentiates anterior ethmoidal artery location on sinus computed tomography scans. J Laryngol Otol. 2020; Jan. 134(1):52–5.
Article21. Chowdhury NI, Smith TL, Chandra RK, Turner JH. Automated classification of osteomeatal complex inflammation on computed tomography using convolutional neural networks. Int Forum Allergy Rhinol. 2019; Jan. 9(1):46–52.
Article22. Wang YM, Li Y, Cheng YS, He ZY, Yang JM, Xu JH, et al. Automated classification of osteomeatal complex inflammation on computed tomography using convolutional neural networks. Int Forum Allergy Rhinol. 2019; Jan. 9(1):46–52.23. Seidler M, Forghani B, Reinhold C, Perez-Lara A, Romero-Sanchez G, Muthukrishnan N, et al. Dual-energy CT texture analysis with machine learning for the evaluation and characterization of cervical lymphadenopathy. Comput Struct Biotechnol J. 2019; Jul. 17:1009–15.
Article24. Hudson TJ, Gare B, Allen DG, Ladak HM, Agrawal SK. Intrinsic measures and shape analysis of the intratemporal facial nerve. Otol Neurotol. 2020; Mar. 41(3):e378–86.
Article25. Kann BH, Hicks DF, Payabvash S, Mahajan A, Du J, Gupta V, et al. Multi-institutional validation of deep learning for pretreatment identification of extranodal extension in head and neck squamous cell carcinoma. J Clin Oncol. 2020; Apr. 38(12):1304–11.
Article26. Parmar C, Grossmann P, Rietveld D, Rietbergen MM, Lambin P, Aerts HJ. Radiomic machine-learning classifiers for prognostic biomarkers of head and neck cancer. Front Oncol. 2015; Dec. 5:272.
Article27. Al Ajmi E, Forghani B, Reinhold C, Bayat M, Forghani R. Spectral multi-energy CT texture analysis with machine learning for tissue classification: an investigation using classification of benign parotid tumours as a testing paradigm. Eur Radiol. 2018; Jun. 28(6):2604–11.
Article28. Tan L, Holland SK, Deshpande AK, Chen Y, Choo DI, Lu LJ. A semi-supervised support vector machine model for predicting the language outcomes following cochlear implantation based on pre-implant brain fMRI imaging. Brain Behav. 2015; Oct. 5(12):e00391.
Article29. Zhang F, Wang JP, Kim J, Parrish T, Wong PC. Decoding multiple sound categories in the human temporal cortex using high resolution fMRI. PLoS One. 2015; Feb. 10(2):e0117303.
Article30. Ergen B, Baykara M, Polat C. Determination of the relationship between internal auditory canal nerves and tinnitus based on the findings of brain magnetic resonance imaging. Biomed Signal Process Control. 2018; Feb. 40:214–9.
Article31. Fujima N, Shimizu Y, Yoshida D, Kano S, Mizumachi T, Homma A, et al. Machine-learning-based prediction of treatment outcomes using MR imaging-derived quantitative tumor information in patients with sinonasal squamous cell carcinomas: a preliminary study. Cancers (Basel). 2019; Jun. 11(6):800.
Article32. Liu Y, Niu H, Zhu J, Zhao P, Yin H, Ding H, et al. Morphological neuroimaging biomarkers for tinnitus: evidence obtained by applying machine learning. Neural Plast. 2019; Dec. 2019:1712342.
Article33. Ramkumar S, Ranjbar S, Ning S, Lal D, Zwart CM, Wood CP, et al. MRI-based texture analysis to differentiate sinonasal squamous cell carcinoma from inverted papilloma. AJNR Am J Neuroradiol. 2017; May. 38(5):1019–25.
Article34. Feng G, Ingvalson EM, Grieco-Calub TM, Roberts MY, Ryan ME, Birmingham P, et al. Neural preservation underlies speech improvement from auditory deprivation in young cochlear implant recipients. Proc Natl Acad Sci U S A. 2018; Jan. 115(5):E1022–31.
Article35. Laves MH, Bicker J, Kahrs LA, Ortmaier T. A dataset of laryngeal endoscopic images with comparative study on convolution neural network-based semantic segmentation. Int J Comput Assist Radiol Surg. 2019; Mar. 14(3):483–92.
Article36. Ren J, Jing X, Wang J, Ren X, Xu Y, Yang Q, et al. Automatic recognition of laryngoscopic images using a deep-learning technique. Laryngoscope. 2020; Feb. 18. [Epub]. https://doi.org/10.1002/lary.28539.
Article37. Xiong H, Lin P, Yu JG, Ye J, Xiao L, Tao Y, et al. Computer-aided diagnosis of laryngeal cancer via deep learning based on laryngoscopic images. EBioMedicine. 2019; Oct. 48:92–9.
Article38. Mascharak S, Baird BJ, Holsinger FC. Detecting oropharyngeal carcinoma using multispectral, narrow-band imaging and machine learning. Laryngoscope. 2018; Nov. 128(11):2514–20.
Article39. Livingstone D, Talai AS, Chau J, Forkert ND. Building an otoscopic screening prototype tool using deep learning. J Otolaryngol Head Neck Surg. 2019; Nov. 48(1):66.
Article40. Tran TT, Fang TY, Pham VT, Lin C, Wang PC, Lo MT. Development of an automatic diagnostic algorithm for pediatric otitis media. Otol Neurotol. 2018; Sep. 39(8):1060–5.
Article41. Livingstone D, Chau J. Otoscopic diagnosis using computer vision: an automated machine learning approach. Laryngoscope. 2020; Jun. 130(6):1408–13.
Article42. Lu C, Lewis JS Jr, Dupont WD, Plummer WD Jr, Janowczyk A, Madabhushi A. An oral cavity squamous cell carcinoma quantitative histomorphometric-based image classifier of nuclear morphology can risk stratify patients for disease-specific survival. Mod Pathol. 2017; Dec. 30(12):1655–65.
Article43. Ashizawa K, Yoshimura K, Johno H, Inoue T, Katoh R, Funayama S, et al. Construction of mass spectra database and diagnosis algorithm for head and neck squamous cell carcinoma. Oral Oncol. 2017; Dec. 75:111–9.
Article44. Grillone GA, Wang Z, Krisciunas GP, Tsai AC, Kannabiran VR, Pistey RW, et al. The color of cancer: margin guidance for oral cancer resection using elastic scattering spectroscopy. Laryngoscope. 2017; Sep. 127 Suppl 4(Suppl 4):S1–9.
Article45. Halicek M, Lu G, Little JV, Wang X, Patel M, Griffith CC, et al. Deep convolutional neural networks for classifying head and neck cancer using hyperspectral imaging. J Biomed Opt. 2017; Jun. 22(6):60503.
Article46. Ma L, Lu G, Wang D, Wang X, Chen ZG, Muller S, et al. Deep learning based classification for head and neck cancer detection with hyperspectral imaging in an animal model. Proc SPIE Int Soc Opt Eng. 2017; Feb. 10137:101372G.
Article47. Lu G, Wang D, Qin X, Muller S, Wang X, Chen AY, et al. Detection and delineation of squamous neoplasia with hyperspectral imaging in a mouse model of tongue carcinogenesis. J Biophotonics. 2018; Mar. 11(3):e201700078.
Article48. Lu G, Little JV, Wang X, Zhang H, Patel MR, Griffith CC, et al. Detection of head and neck cancer in surgical specimens using quantitative hyperspectral imaging. Clin Cancer Res. 2017; Sep. 23(18):5426–36.
Article49. Lin J, Clancy NT, Qi J, Hu Y, Tatla T, Stoyanov D, et al. Dual-modality endoscopic probe for tissue surface shape reconstruction and hyperspectral imaging enabled by deep neural networks. Med Image Anal. 2018; Aug. 48:162–76.
Article50. Halicek M, Dormer JD, Little JV, Chen AY, Myers L, Sumer BD, et al. Hyperspectral imaging of head and neck squamous cell carcinoma for cancer margin detection in surgical specimens from 102 patients using deep learning. Cancers (Basel). 2019; Sep. 11(9):1367.
Article51. Fei B, Lu G, Wang X, Zhang H, Little JV, Patel MR, et al. Label-free reflectance hyperspectral imaging for tumor margin assessment: a pilot study on surgical specimens of cancer patients. J Biomed Opt. 2017; Aug. 22(8):1–7.
Article52. Halicek M, Little JV, Wang X, Chen AY, Fei B. Optical biopsy of head and neck cancer using hyperspectral imaging and convolutional neural networks. J Biomed Opt. 2019; Mar. 24(3):1–9.
Article53. Zhang L, Wu Y, Zheng B, Su L, Chen Y, Ma S, et al. Rapid histology of laryngeal squamous cell carcinoma with deep-learning based stimulated Raman scattering microscopy. Theranostics. 2019; Apr. 9(9):2541–54.
Article54. Halicek M, Little JV, Wang X, Patel M, Griffith CC, Chen AY, et al. Tumor margin classification of head and neck cancer using hyperspectral imaging and convolutional neural networks. Proc SPIE Int Soc Opt Eng. 2018; Feb. 10576:1057605.
Article55. Daniels K, Gummadi S, Zhu Z, Wang S, Patel J, Swendseid B, et al. Machine learning by ultrasonography for genetic risk stratification of thyroid nodules. JAMA Otolaryngol Head Neck Surg. 2019; Oct. 146(1):1–6.
Article56. Parmar P, Habib AR, Mendis D, Daniel A, Duvnjak M, Ho J, et al. An artificial intelligence algorithm that identifies middle turbinate pneumatisation (concha bullosa) on sinus computed tomography scans. J Laryngol Otol. 2020; Apr. 134(4):328–31.
Article57. Murata M, Ariji Y, Ohashi Y, Kawai T, Fukuda M, Funakoshi T, et al. Deep-learning classification using convolutional neural network for evaluation of maxillary sinusitis on panoramic radiography. Oral Radiol. 2019; Sep. 35(3):301–7.
Article58. Lai YH, Tsao Y, Lu X, Chen F, Su YT, Chen KC, et al. Deep learning-based noise reduction approach to improve speech intelligibility for cochlear implant recipients. Ear Hear. 2018; Jul/Aug. 39(4):795–809.
Article59. Healy EW, Yoho SE, Chen J, Wang Y, Wang D. An algorithm to increase speech intelligibility for hearing-impaired listeners in novel segments of the same noise type. J Acoust Soc Am. 2015; Sep. 138(3):1660–9.
Article60. Erfanian Saeedi N, Blamey PJ, Burkitt AN, Grayden DB. An integrated model of pitch perception incorporating place and temporal pitch codes with application to cochlear implant research. Hear Res. 2017; Feb. 344:135–47.
Article61. Guerra-Jimenez G, Ramos De Miguel A, Falcon Gonzalez JC, Borkoski Barreiro SA, Perez Plasencia D, Ramos Macias A. Cochlear implant evaluation: prognosis estimation by data mining system. J Int Adv Otol. 2016; Apr. 12(1):1–7.62. Lai YH, Chen F, Wang SS, Lu X, Tsao Y, Lee CH. A deep denoising autoencoder approach to improving the intelligibility of vocoded speech in cochlear implant simulation. IEEE Trans Biomed Eng. 2017; Jul. 64(7):1568–78.
Article63. Chen J, Wang Y, Yoho SE, Wang D, Healy EW. Large-scale training to increase speech intelligibility for hearing-impaired listeners in novel noises. J Acoust Soc Am. 2016; May. 139(5):2604.
Article64. Gao X, Grayden DB, McDonnell MD. Modeling electrode place discrimination in cochlear implant stimulation. IEEE Trans Biomed Eng. 2017; Sep. 64(9):2219–29.
Article65. Hajiaghababa F, Marateb HR, Kermani S. The design and validation of a hybrid digital-signal-processing plug-in for traditional cochlear implant speech processors. Comput Methods Programs Biomed. 2018; Jun. 159:103–9.
Article66. Ramos-Miguel A, Perez-Zaballos T, Perez D, Falconb JC, Ramosb A. Use of data mining to predict significant factors and benefits of bilateral cochlear implantation. Eur Arch Otorhinolaryngol. 2015; Nov. 272(11):3157–62.
Article67. Powell ME, Rodriguez Cancio M, Young D, Nock W, Abdelmessih B, Zeller A, et al. Decoding phonation with artificial intelligence (DeP AI): proof of concept. Laryngoscope Investig Otolaryngol. 2019; Mar. 4(3):328–34.68. Tsui SY, Tsao Y, Lin CW, Fang SH, Lin FC, Wang CT. Demographic and symptomatic features of voice disorders and their potential application in classification using machine learning algorithms. Folia Phoniatr Logop. 2018; 70(3-4):174–82.
Article69. Fang SH, Tsao Y, Hsiao MJ, Chen JY, Lai YH, Lin FC, et al. Detection of pathological voice using cepstrum vectors: a deep learning approach. J Voice. 2019; Sep. 33(5):634–41.
Article70. Fujimura S, Kojima T, Okanoue Y, Shoji K, Inoue M, Hori R. Discrimination of “hot potato voice” caused by upper airway obstruction utilizing a support vector machine. Laryngoscope. 2019; Jun. 129(6):1301–7.
Article71. Rameau A. Pilot study for a novel and personalized voice restoration device for patients with laryngectomy. Head Neck. 2020; May. 42(5):839–45.
Article72. Zhang L, Fabbri D, Upender R, Kent D. Automated sleep stage scoring of the Sleep Heart Health Study using deep neural networks. Sleep. 2019; Oct. 42(11):zsz159.
Article73. Zhang X, Xu M, Li Y, Su M, Xu Z, Wang C, et al. Automated multi-model deep neural network for sleep stage scoring with unfiltered clinical data. Sleep Breath. 2020; Jun. 24(2):581–90.
Article74. Zhang L, Wu H, Zhang X, Wei X, Hou F, Ma Y. Sleep heart rate variability assists the automatic prediction of long-term cardiovascular outcomes. Sleep Med. 2020; Mar. 67:217–24.
Article75. Yao J, Zhang YT. The application of bionic wavelet transform to speech signal processing in cochlear implants using neural network simulations. IEEE Trans Biomed Eng. 2002; Nov. 49(11):1299–309.76. Nemati P, Imani M, Farahmandghavi F, Mirzadeh H, Marzban-Rad E, Nasrabadi AM. Artificial neural networks for bilateral prediction of formulation parameters and drug release profiles from cochlear implant coatings fabricated as porous monolithic devices based on silicone rubber. J Pharm Pharmacol. 2014; May. 66(5):624–38.
Article77. Middlebrooks JC, Bierer JA. Auditory cortical images of cochlear-implant stimuli: coding of stimulus channel and current level. J Neurophysiol. 2002; Jan. 87(1):493–507.
Article78. Charasse B, Thai-Van H, Chanal JM, Berger-Vachon C, Collet L. Automatic analysis of auditory nerve electrically evoked compound action potential with an artificial neural network. Artif Intell Med. 2004; Jul. 31(3):221–9.
Article79. Botros A, van Dijk B, Killian M. AutoNR: an automated system that measures ECAP thresholds with the Nucleus Freedom cochlear implant via machine intelligence. Artif Intell Med. 2007; May. 40(1):15–28.80. van Dijk B, Botros AM, Battmer RD, Begall K, Dillier N, Hey M, et al. Clinical results of AutoNRT, a completely automatic ECAP recording system for cochlear implants. Ear Hear. 2007; Aug. 28(4):558–70.
Article81. Gartner L, Lenarz T, Joseph G, Buchner A. Clinical use of a system for the automated recording and analysis of electrically evoked compound action potentials (ECAPs) in cochlear implant patients. Acta Otolaryngol. 2010; Jun. 130(6):724–32.82. Nemati P, Imani M, Farahmandghavi F, Mirzadeh H, Marzban-Rad E, Nasrabadi AM. Dexamethasone-releasing cochlear implant coatings: application of artificial neural networks for modelling of formulation parameters and drug release profile. J Pharm Pharmacol. 2013; Aug. 65(8):1145–57.
Article83. Zhang J, Wei W, Ding J, Roland JT Jr, Manolidis S, Simaan N. Inroads toward robot-assisted cochlear implant surgery using steerable electrode arrays. Otol Neurotol. 2010; Oct. 31(8):1199–206.
Article84. Chang CH, Anderson GT, Loizou PC. A neural network model for optimizing vowel recognition by cochlear implant listeners. IEEE Trans Neural Syst Rehabil Eng. 2001; Mar. 9(1):42–8.85. Castaneda-Villa N, James CJ. Objective source selection in blind source separation of AEPs in children with cochlear implants. Conf Proc IEEE Eng Med Biol Soc. 2007; 2007:6224–7.
Article86. Desmond JM, Collins LM, Throckmorton CS. Using channel-specific statistical models to detect reverberation in cochlear implant stimuli. J Acoust Soc Am. 2013; Aug. 134(2):1112–20.
Article87. Kim JW, Kim T, Shin J, Lee K, Choi S, Cho SW. Prediction of apnea-hypopnea index using sound data collected by a noncontact device. Otolaryngol Head Neck Surg. 2020; Mar. 162(3):392–9.
Article88. Ruiz EM, Niu T, Zerfaoui M, Kunnimalaiyaan M, Friedlander PL, Abdel-Mageed AB, et al. A novel gene panel for prediction of lymphnode metastasis and recurrence in patients with thyroid cancer. Surgery. 2020; Jan. 167(1):73–9.
Article89. Zhong Q, Fang J, Huang Z, Yang Y, Lian M, Liu H, et al. A response prediction model for taxane, cisplatin, and 5-fluorouracil chemotherapy in hypopharyngeal carcinoma. Sci Rep. 2018; Aug. 8(1):12675.
Article90. Chowdhury NI, Li P, Chandra RK, Turner JH. Baseline mucus cytokines predict 22-item Sino-Nasal Outcome Test results after endoscopic sinus surgery. Int Forum Allergy Rhinol. 2020; Jan. 10(1):15–22.
Article91. Urata S, Iida T, Yamamoto M, Mizushima Y, Fujimoto C, Matsumoto Y, et al. Cellular cartography of the organ of Corti based on optical tissue clearing and machine learning. Elife. 2019; Jan. 8:e40946.
Article92. Zhao Z, Li Y, Wu Y, Chen R. Deep learning-based model for predicting progression in patients with head and neck squamous cell carcinoma. Cancer Biomark. 2020; 27(1):19–28.
Article93. Essers PBM, van der Heijden M, Verhagen CV, Ploeg EM, de Roest RH, Leemans CR, et al. Drug sensitivity prediction models reveal a link between DNA repair defects and poor prognosis in HNSCC. Cancer Res. 2019; Nov. 79(21):5597–611.
Article94. Ishii H, Saitoh M, Sakamoto K, Sakamoto K, Saigusa D, Kasai H, et al. Lipidome-based rapid diagnosis with machine learning for detection of TGF-β signalling activated area in head and neck cancer. Br J Cancer. 2020; Mar. 122(7):995–1004.
Article95. Patel KN, Angell TE, Babiarz J, Barth NM, Blevins T, Duh QY, et al. Performance of a genomic sequencing classifier for the preoperative diagnosis of cytologically indeterminate thyroid nodules. JAMA Surg. 2018; Sep. 153(9):817–24.
Article96. Stepp WH, Farquhar D, Sheth S, Mazul A, Mamdani M, Hackman TG, et al. RNA oncoimmune phenotyping of HPV-positive p16-positive oropharyngeal squamous cell carcinomas by nodal status. Version 2. JAMA Otolaryngol Head Neck Surg. 2018; Nov. 144(11):967–75.97. Shew M, New J, Wichova H, Koestler DC, Staecker H. Using machine learning to predict sensorineural hearing loss based on perilymph micro RNA expression profile. Sci Rep. 2019; Mar. 9(1):3393.
Article98. Nam Y, Choo OS, Lee YR, Choung YH, Shin H. Cascade recurring deep networks for audible range prediction. BMC Med Inform Decis Mak. 2017; May. 17(Suppl 1):56.
Article99. Kim H, Kang WS, Park HJ, Lee JY, Park JW, Kim Y, et al. Cochlear implantation in postlingually deaf adults is time-sensitive towards positive outcome: prediction using advanced machine learning techniques. Sci Rep. 2018; Dec. 8(1):18004.
Article100. Bing D, Ying J, Miao J, Lan L, Wang D, Zhao L, et al. Predicting the hearing outcome in sudden sensorineural hearing loss via machine learning models. Clin Otolaryngol. 2018; Jun. 43(3):868–74.
Article101. Lau K, Wilkinson J, Moorthy R. A web-based prediction score for head and neck cancer referrals. Clin Otolaryngol. 2018; Mar. 43(4):1043–9.
Article102. Wilson MB, Ali SA, Kovatch KJ, Smith JD, Hoff PT. Machine learning diagnosis of peritonsillar abscess. Otolaryngol Head Neck Surg. 2019; Nov. 161(5):796–9.
Article103. Priesol AJ, Cao M, Brodley CE, Lewis RF. Clinical vestibular testing assessed with machine-learning algorithms. JAMA Otolaryngol Head Neck Surg. 2015; Apr. 141(4):364–72.
Article104. Chan J, Raju S, Nandakumar R, Bly R, Gollakota S. Detecting middle ear fluid using smartphones. Sci Transl Med. 2019; May. 11(492):eaav1102.
Article105. Karadaghy OA, Shew M, New J, Bur AM. Development and assessment of a machine learning model to help predict survival among patients with oral squamous cell carcinoma. JAMA Otolaryngol Head Neck Surg. 2019; May. 145(12):1115–20.
Article106. Mermod M, Jourdan EF, Gupta R, Bongiovanni M, Tolstonog G, Simon C, et al. Development and validation of a multivariable prediction model for the identification of occult lymph node metastasis in oral squamous cell carcinoma. Head Neck. 2020; Aug. 42(8):1811–20.
Article107. Formeister EJ, Baum R, Knott PD, Seth R, Ha P, Ryan W, et al. Machine learning for predicting complications in head and neck microvascular free tissue transfer. Laryngoscope. 2020; Jan. 28. [Epub]. https://doi.org/10.1002/lary.28508.
Article108. Shew M, New J, Bur AM. Machine learning to predict delays in adjuvant radiation following surgery for head and neck cancer. Otolaryngol Head Neck Surg. 2019; Jun. 160(6):1058–64.
Article109. Bur AM, Holcomb A, Goodwin S, Woodroof J, Karadaghy O, Shnayder Y, et al. Machine learning to predict occult nodal metastasis in early oral squamous cell carcinoma. Oral Oncol. 2019; May. 92:20–5.
Article110. Kamogashira T, Fujimoto C, Kinoshita M, Kikkawa Y, Yamasoba T, Iwasaki S. Prediction of vestibular dysfunction by applying machine learning algorithms to postural instability. Front Neurol. 2020; Feb. 11:7.
Article111. Liu YW, Kao SL, Wu HT, Liu TC, Fang TY, Wang PC. Transient-evoked otoacoustic emission signals predicting outcomes of acute sensorineural hearing loss in patients with Meniere’s disease. Acta Otolaryngol. 2020; Mar. 140(3):230–5.112. Luo J, Erbe C, Friedland DR. Unique clinical language patterns among expert vestibular providers can predict vestibular diagnoses. Otol Neurotol. 2018; Oct. 39(9):1163–71.
Article113. Wu Q, Chen J, Deng H, Ren Y, Sun Y, Wang W, et al. Expert-level diagnosis of nasal polyps using deep learning on whole-slide imaging. J Allergy Clin Immunol. 2020; Feb. 145(2):698–701.
Article114. Jabez Christopher J, Khanna Nehemiah H, Kannan A. A clinical decision support system for diagnosis of allergic rhinitis based on intradermal skin tests. Comput Biol Med. 2015; Oct. 65:76–84.
Article115. Adnane C, Adouly T, Khallouk A, Rouadi S, Abada R, Roubal M, et al. Using preoperative unsupervised cluster analysis of chronic rhinosinusitis to inform patient decision and endoscopic sinus surgery outcome. Eur Arch Otorhinolaryngol. 2017; Feb. 274(2):879–85.
Article116. Soler ZM, Hyer JM, Rudmik L, Ramakrishnan V, Smith TL, Schlosser RJ. Cluster analysis and prediction of treatment outcomes for chronic rhinosinusitis. J Allergy Clin Immunol. 2016; Apr. 137(4):1054–62.
Article117. Soler ZM, Hyer JM, Ramakrishnan V, Smith TL, Mace J, Rudmik L, et al. Identification of chronic rhinosinusitis phenotypes using cluster analysis. Int Forum Allergy Rhinol. 2015; May. 5(5):399–407.
Article118. Divekar R, Patel N, Jin J, Hagan J, Rank M, Lal D, et al. Symptom-based clustering in chronic rhinosinusitis relates to history of aspirin sensitivity and postsurgical outcomes. J Allergy Clin Immunol Pract. 2015; Nov-Dec. 3(6):934–40.
Article119. Lal D, Hopkins C, Divekar RD. SNOT-22-based clusters in chronic rhinosinusitis without nasal polyposis exhibit distinct endotypic and prognostic differences. Int Forum Allergy Rhinol. 2018; Jul. 8(7):797–805.
Article120. Tomassen P, Vandeplas G, Van Zele T, Cardell LO, Arebro J, Olze H, et al. Inflammatory endotypes of chronic rhinosinusitis based on cluster analysis of biomarkers. J Allergy Clin Immunol. 2016; May. 137(5):1449–56.
Article121. Kim JW, Huh G, Rhee CS, Lee CH, Lee J, Chung JH, et al. Unsupervised cluster analysis of chronic rhinosinusitis with nasal polyp using routinely available clinical markers and its implication in treatment outcomes. Int Forum Allergy Rhinol. 2019; Jan. 9(1):79–86.
Article122. Goldstein CA, Berry RB, Kent DT, Kristo DA, Seixas AA, Redline S, et al. Artificial intelligence in sleep medicine: background and implications for clinicians. J Clin Sleep Med. 2020; Apr. 16(4):609–18.
Article123. Weininger O, Warnecke A, Lesinski-Schiedat A, Lenarz T, Stolle S. Computational analysis based on audioprofiles: a new possibility for patient stratification in office-based otology. Audiol Res. 2019; Nov. 9(2):230.
Article124. Barbour DL, Howard RT, Song XD, Metzger N, Sukesan KA, DiLorenzo JC, et al. Online machine learning audiometry. Ear Hear. 2019; Jul/Aug. 40(918):26.
Article125. Wu YH, Ho HC, Hsiao SH, Brummet RB, Chipara O. Predicting three-month and 12-month post-fitting real-world hearing-aid outcome using pre-fitting acceptable noise level (ANL). Int J Audiol. 2016; 55(5):285–94.
Article126. Bramhall NF, McMillan GP, Kujawa SG, Konrad-Martin D. Use of non-invasive measures to predict cochlear synapse counts. Hear Res. 2018; Dec. 370:113–9.
Article127. Rasku J, Pyykko I, Levo H, Kentala E, Manchaiah V. Disease profiling for computerized peer support of Meniere’s disease. JMIR Rehabil Assist Technol. 2015; Sep. 2(2):e9.128. Morse JC, Shilts MH, Ely KA, Li P, Sheng Q, Huang LC, et al. Patterns of olfactory dysfunction in chronic rhinosinusitis identified by hierarchical cluster analysis and machine learning algorithms. Int Forum Allergy Rhinol. 2019; Mar. 9(3):255–64.
Article129. Quon H, Hui X, Cheng Z, Robertson S, Peng L, Bowers M, et al. Quantitative evaluation of head and neck cancer treatment-related dysphagia in the development of a personalized treatment deintensification paradigm. Int J Radiat Oncol Biol Phys. 2017; Dec. 99(5):1271–8.
Article130. Jochems A, Leijenaar RT, Bogowicz M, Hoebers FJ, Wesseling F, Huang SH, et al. Combining deep learning and radiomics to predict HPV status in oropharyngeal squamous cell carcinoma. Radiat Oncol. 2018; Apr. 127:S504–5.131. Bao T, Klatt BN, Whitney SL, Sienko KH, Wiens J. Automatically evaluating balance: a machine learning approach. IEEE Trans Neural Syst Rehabil Eng. 2019; Feb. 27(2):179–86.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Current Developments of Artificial Intelligences in Head and Neck Cancer Histopathological Images Analysis
- Artificial intelligence in obstetrics
- Artificial Intelligence in Pathology
- Application of Machine Learning in Rhinology: A State of the Art Review
- Research and Publication Ethics in Korean Journal of Otorhinolaryngology-Head and Neck Surgery