Cancer Res Treat.
2020 Oct;52(4):1273-1282. 10.4143/crt.2020.032.
Forkhead Box C1 (FOXC1) Expression in Stromal Cells within the Microenvironment of T and NK Cell Lymphomas: Association with Tumor Dormancy and Activation
- Affiliations
-
- 1Department of Pathology, Severance Hospital, Yonsei University College of Medicine, Seoul, Korea
- KMID: 2507952
- DOI: http://doi.org/10.4143/crt.2020.032
Abstract
- Purpose
Forkhead box C1 (FOXC1) is critical for maintaining bone marrow microenvironments during hematopoiesis, but its role in hematological malignancies remains obscure. Here, we investigated whether FOXC1 regulates tumor dormancy and activation in the microenvironments of T and natural killer (NK) cell lymphomas.
Materials and Methods
One hundred and twenty cases of T and NK cell lymphomas were included; the immunohistochemical expression of FOXC1 was investigated in stromal cells, and numbers of FOXC1+ stromal cells were counted. Furthermore, the expression of phosphorylated p38 (p-p38) and phosphorylated ERK1/2 (p-ERK1/2) in tumor cells was investigated using immunohistochemistry.
Results
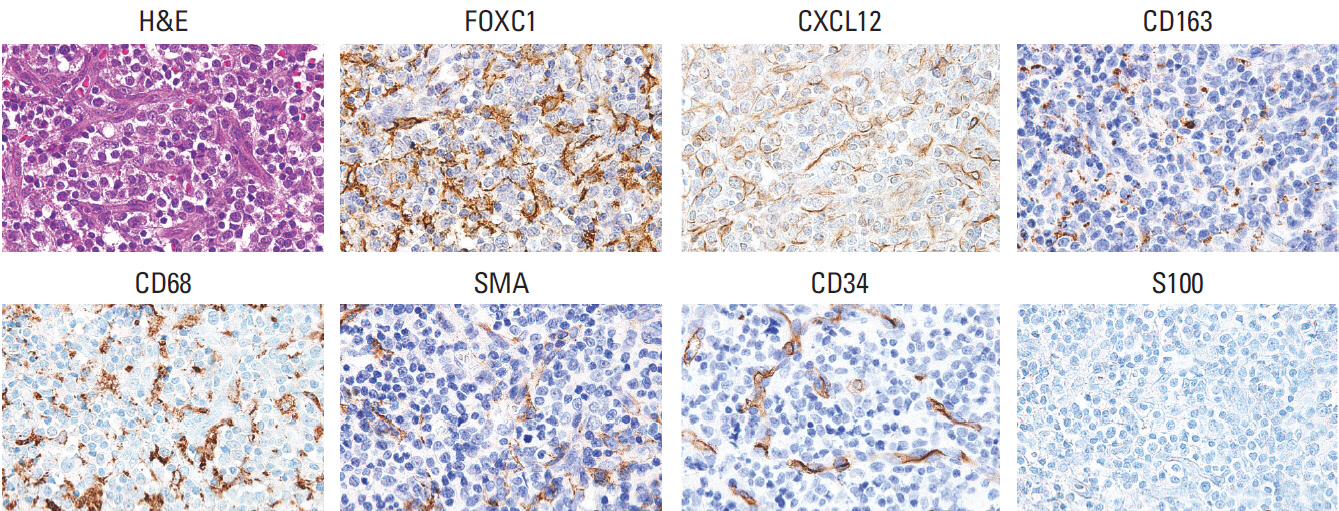
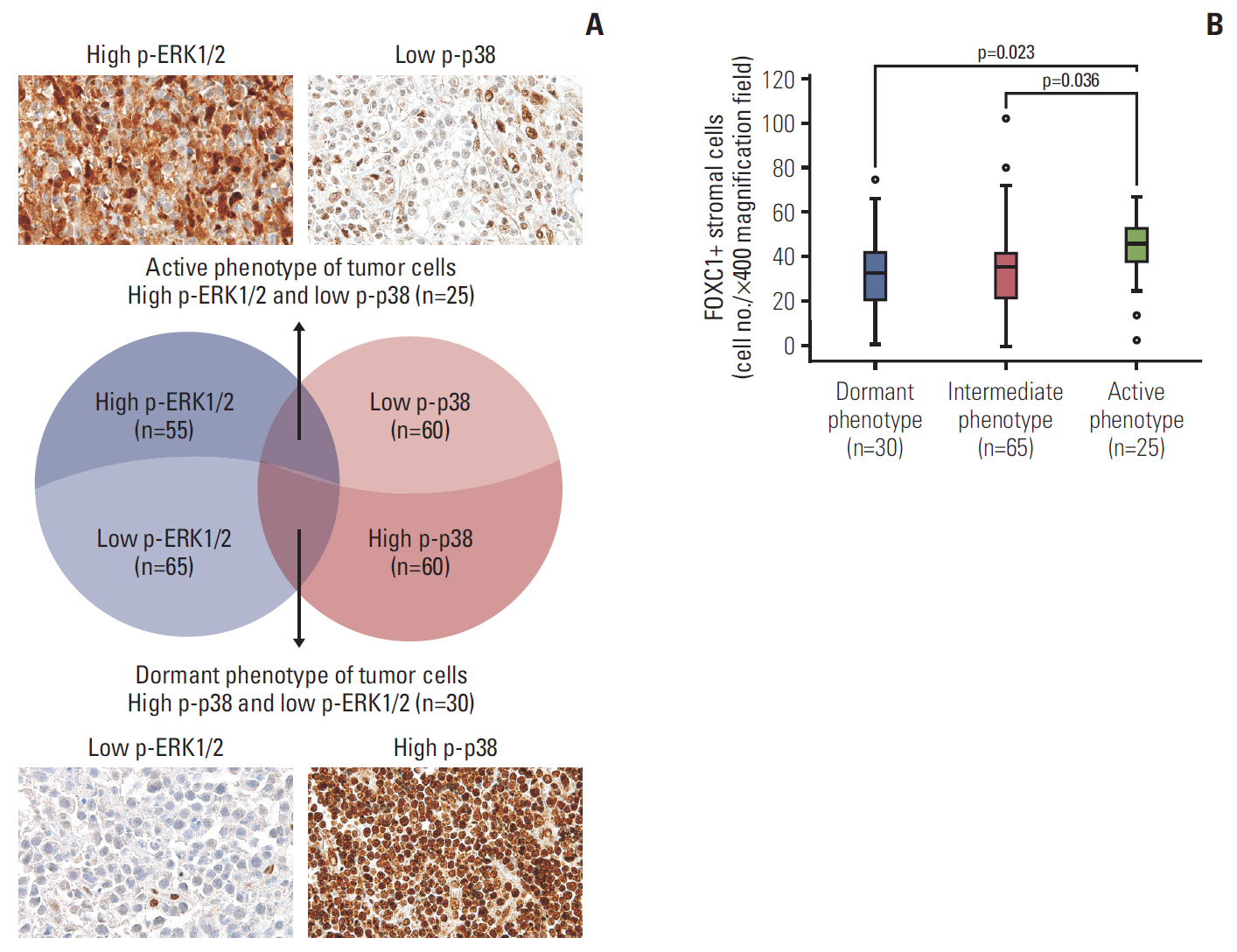
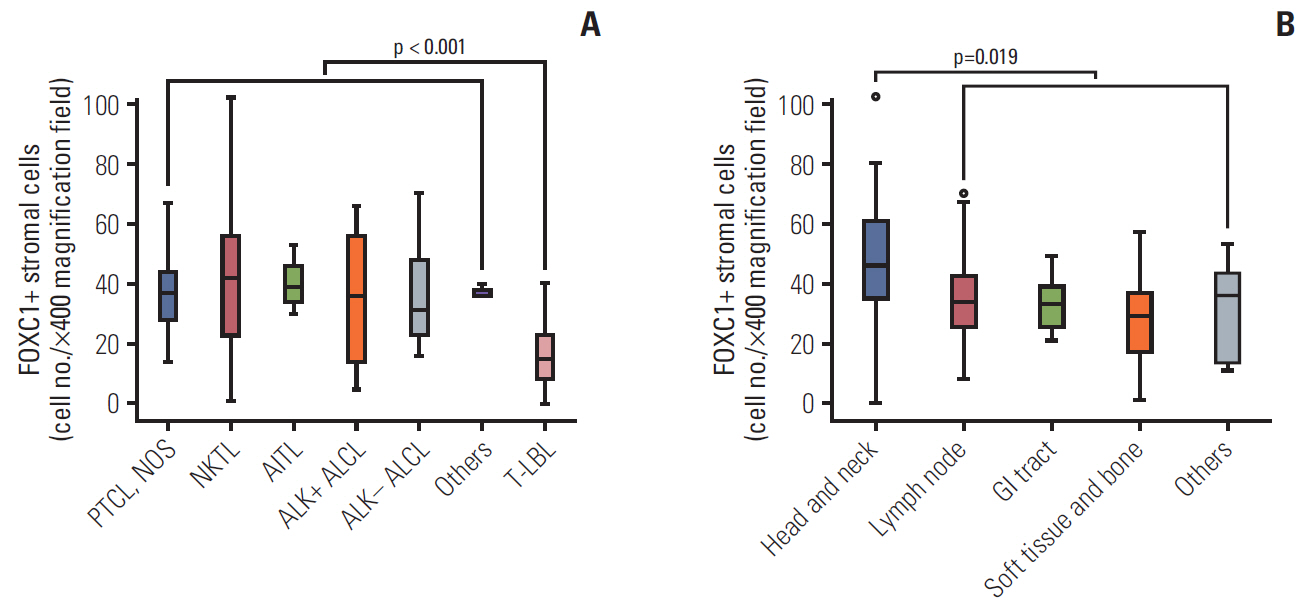
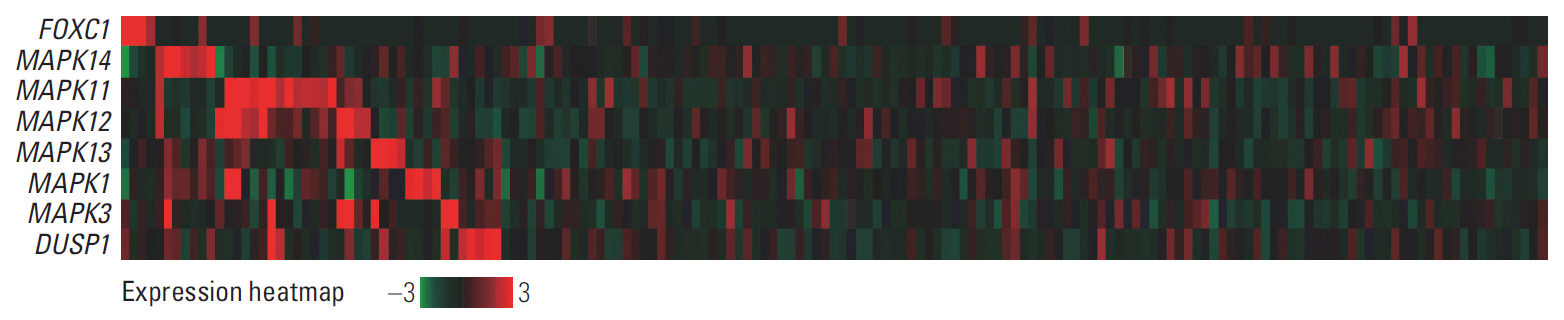
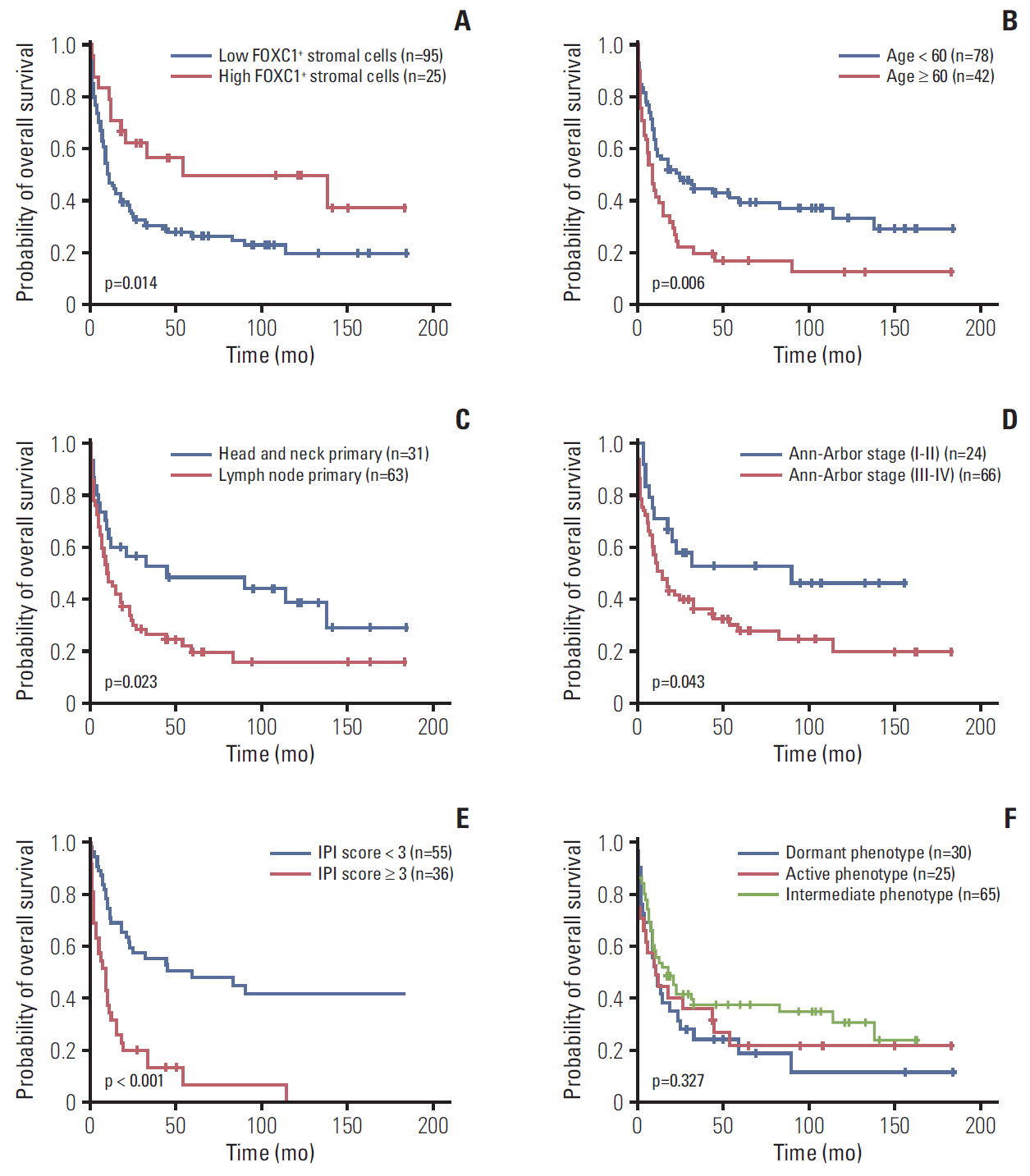
FOXC1 was variably expressed in C-X-C motif chemokine 12–associated reticular stromal cells, histiocytes, (myo)fibroblasts, and endothelial cells. The phenotypes of cases were categorized as dormant (high p-p38/low p-ERK1/2; n=30, 25.0%), active (high p-ERK1/2/low p-p38; n=25, 20.8%), or intermediate (others; n=65, 54.2%). Lower FOXC1+ stromal cell infiltration was associated with the dormant phenotype, the precursor T lymphoblastic leukemia/lymphoma subtype, and inferior overall survival rates, whereas higher FOXC1+ stromal cell infiltration was associated with the active phenotype and favorable patient prognosis (p < 0.05 for all).
Conclusion
These results suggested that FOXC1+ stromal cells within the microenvironments of T and NK cell lymphomas might be related to tumor phenotypes.
Keyword
Figure
Reference
-
References
1. Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013; 19:1423–37.
Article2. Yumoto K, Eber MR, Berry JE, Taichman RS, Shiozawa Y. Molecular pathways: niches in metastatic dormancy. Clin Cancer Res. 2014; 20:3384–9.
Article3. Sosa MS, Bragado P, Aguirre-Ghiso JA. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat Rev Cancer. 2014; 14:611–22.
Article4. Lehmann OJ, Sowden JC, Carlsson P, Jordan T, Bhattacharya SS. Fox’s in development and disease. Trends Genet. 2003; 19:339–44.
Article5. Hannenhalli S, Kaestner KH. The evolution of Fox genes and their role in development and disease. Nat Rev Genet. 2009; 10:233–40.
Article6. Nagel S, Meyer C, Kaufmann M, Drexler HG, MacLeod RA. Deregulated FOX genes in Hodgkin lymphoma. Genes Chromosomes Cancer. 2014; 53:917–33.
Article7. Liu J, Shen L, Yao J, Li Y, Wang Y, Chen H, et al. Forkhead box C1 promoter upstream transcript, a novel long non-coding RNA, regulates proliferation and migration in basal-like breast cancer. Mol Med Rep. 2015; 11:3155–9.
Article8. Wang J, Li L, Liu S, Zhao Y, Wang L, Du G. FOXC1 promotes melanoma by activating MST1R/PI3K/AKT. Oncotarget. 2016; 7:84375–87.9. Omatsu Y, Seike M, Sugiyama T, Kume T, Nagasawa T. Foxc1 is a critical regulator of haematopoietic stem/progenitor cell niche formation. Nature. 2014; 508:536–40.
Article10. Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006; 25:977–88.
Article11. Greenbaum A, Hsu YM, Day RB, Schuettpelz LG, Christopher MJ, Borgerding JN, et al. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature. 2013; 495:227–30.
Article12. Omatsu Y, Nagasawa T. The critical and specific transcriptional regulator of the microenvironmental niche for hematopoietic stem and progenitor cells. Curr Opin Hematol. 2015; 22:330–6.
Article13. Diehl NL, Enslen H, Fortner KA, Merritt C, Stetson N, Charland C, et al. Activation of the p38 mitogen-activated protein kinase pathway arrests cell cycle progression and differentiation of immature thymocytes in vivo. J Exp Med. 2000; 191:321–34.
Article14. Crompton T, Gilmour KC, Owen MJ. The MAP kinase pathway controls differentiation from double-negative to double-positive thymocyte. Cell. 1996; 86:243–51.
Article15. Vose J, Armitage J; Weisenburger; International T-Cell Lymphoma Project. International peripheral T-cell and natural killer/T-cell lymphoma study: pathology findings and clinical outcomes. J Clin Oncol. 2008; 26:4124–30.16. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. WHO classification of tumours of haematopoietic and lymphoid tissues. 4th ed. Lyon: International Agency for Research on Cancer;2008.17. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. WHO classification of tumours of haematopoietic and lymphoid tissues. Revised 4th ed. Lyon: International Agency for Research on Cancer;2017.18. Kim SH, Yang WI, Min YH, Ko YH, Yoon SO. The role of the polycomb repressive complex pathway in T and NK cell lymphoma: biological and prognostic implications. Tumour Biol. 2016; 37:2037–47.
Article19. Dave SS, Wright G, Tan B, Rosenwald A, Gascoyne RD, Chan WC, et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med. 2004; 351:2159–69.20. Lenz G, Wright G, Dave SS, Xiao W, Powell J, Zhao H, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008; 359:2313–23.
Article21. Masiero M, Minuzzo S, Pusceddu I, Moserle L, Persano L, Agnusdei V, et al. Notch3-mediated regulation of MKP-1 levels promotes survival of T acute lymphoblastic leukemia cells. Leukemia. 2011; 25:588–98.
Article22. Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Cancer. 2007; 7:834–46.
Article23. Kumar V, Abbas AK, Aster JC. Robbins and Cotran pathologic basis of disease. 9th ed. Philadelphia, PA: Elsevier/Saunders;2015.24. Ray PS, Bagaria SP, Wang J, Shamonki JM, Ye X, Sim MS, et al. Basal-like breast cancer defined by FOXC1 expression offers superior prognostic value: a retrospective immunohistochemical study. Ann Surg Oncol. 2011; 18:3839–47.
Article25. Yamaguchi H, Hung MC. Regulation and role of EZH2 in cancer. Cancer Res Treat. 2014; 46:209–22.
Article26. Xu Y, Shao QS, Yao HB, Jin Y, Ma YY, Jia LH. Overexpression of FOXC1 correlates with poor prognosis in gastric cancer patients. Histopathology. 2014; 64:963–70.
Article27. Wei LX, Zhou RS, Xu HF, Wang JY, Yuan MH. High expression of FOXC1 is associated with poor clinical outcome in non-small cell lung cancer patients. Tumour Biol. 2013; 34:941–6.
Article28. Liu Z, Xu S, Chu H, Lu Y, Yuan P, Zeng X. Silencing FOXC1 inhibits growth and migration of human oral squamous cell carcinoma cells. Exp Ther Med. 2018; 16:3369–76.
Article29. Berry FB, Saleem RA, Walter MA. FOXC1 transcriptional regulation is mediated by N- and C-terminal activation domains and contains a phosphorylated transcriptional inhibitory domain. J Biol Chem. 2002; 277:10292–7.
Article30. Berry FB, Mirzayans F, Walter MA. Regulation of FOXC1 stability and transcriptional activity by an epidermal growth factor-activated mitogen-activated protein kinase signaling cascade. J Biol Chem. 2006; 281:10098–104.
Article31. Wang LY, Li LS, Yang Z. Correlation of FOXC1 protein with clinicopathological features in serous ovarian tumors. Oncol Lett. 2016; 11:933–8.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Upregulated Neuro-oncological Ventral Antigen 1 (NOVA1) Expression Is Specific to Mature and Immature T- and NK-Cell Lymphomas
- Cellular Dormancy in Cancer: Mechanisms and Potential Targeting Strategies
- Granzyme B immunoreactivity in T/natural killer cell lymphomas
- Immuno-oncology for B-cell lymphomas
- Exosomal Communication Between the Tumor Microenvironment and Innate Immunity and Its Therapeutic Application