Cancer Res Treat.
2020 Jul;52(3):697-713. 10.4143/crt.2019.559.
Detection of Germline Mutations in Breast Cancer Patients with Clinical Features of Hereditary Cancer Syndrome Using a Multi-Gene Panel Test
- Affiliations
-
- 1Department of Surgery, Seoul National University Bundang Hospital, Seongnam, Korea
- 2Department of Surgery, Seoul National University Hospital, Seoul, Korea
- 3Department of Surgery, Seoul St. Mary's Hospital, College of Medicine, Seoul, Korea
- 4Center for Medical Innovation, Biomedical Research Institute, Seoul National University Hospital, Seoul, Korea
- 5National Cancer Center-Graduate School for Cancer Science and Policy, Goyang, Korea
- 6College of Veterinary Medicine, Konkuk University, Seoul, Korea
- 7Clinical Genomics Analysis Branch, Research Institute, National Cancer Center, Goyang, Korea
- 8Center for Breast Cancer, Hospital, National Cancer Center, Goyang, Korea
- 9Translational Cancer Research Branch, Division of Translational Science, National Cancer Center, Goyang, Korea
- 10Cancer Research Institute, Seoul National University, Seoul, Korea
- 11Genetic Counseling Clinic, Hospital, Department of System Cancer Science, Graduate School of Cancer Science and Policy, Goyang, Korea
- KMID: 2504452
- DOI: http://doi.org/10.4143/crt.2019.559
Abstract
- Purpose
Hereditary cancer syndrome means that inherited genetic mutations can increase a person's risk of developing cancer. We assessed the frequency of germline mutations using an nextgeneration sequencing (NGS)–based multiple-gene panel containing 64 cancer-predisposing genes in Korean breast cancer patients with clinical features of hereditary breast and ovarian cancer syndrome (HBOC).
Materials and Methods
A total of 64 genes associated with hereditary cancer syndrome were selected for development of an NGS-based multi-gene panel. Targeted sequencing using the multi-gene panel was performed to identify germline mutations in 496 breast cancer patients with clinical features of HBOC who underwent breast cancer surgery between January 2002 and December 2017.
Results
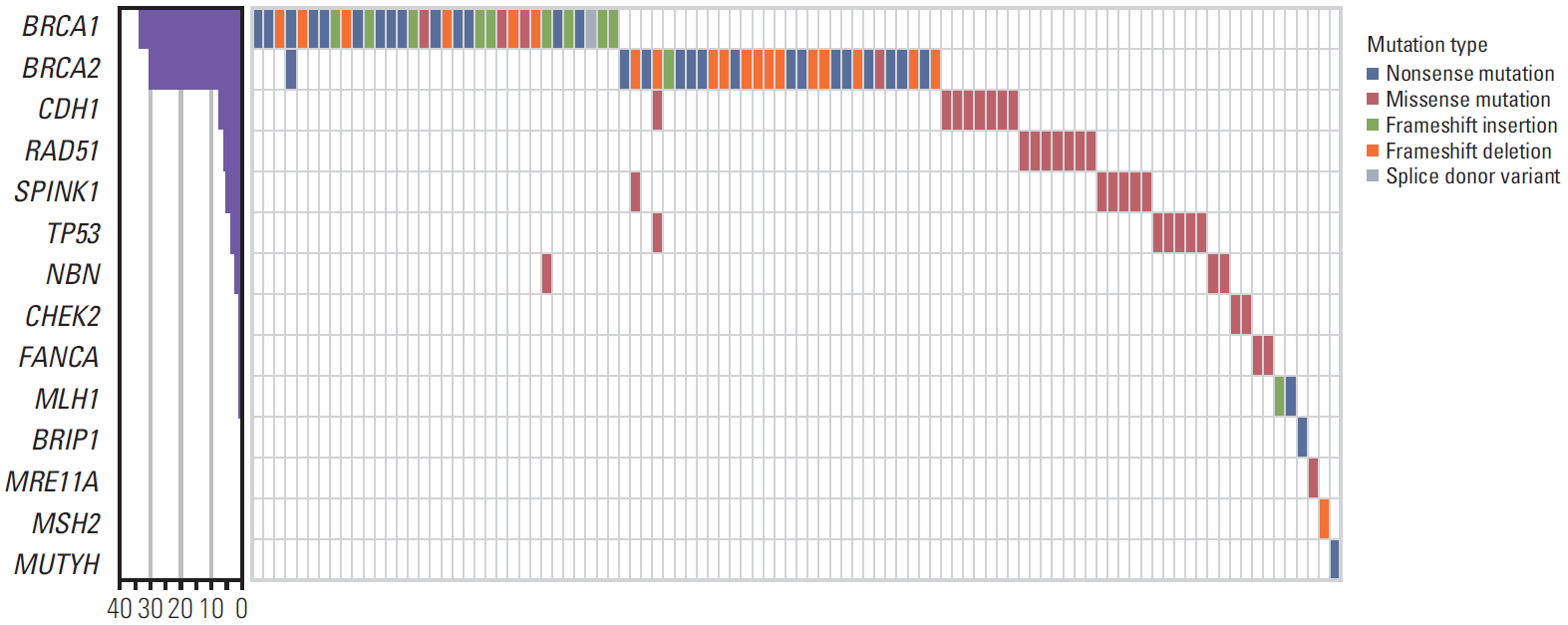
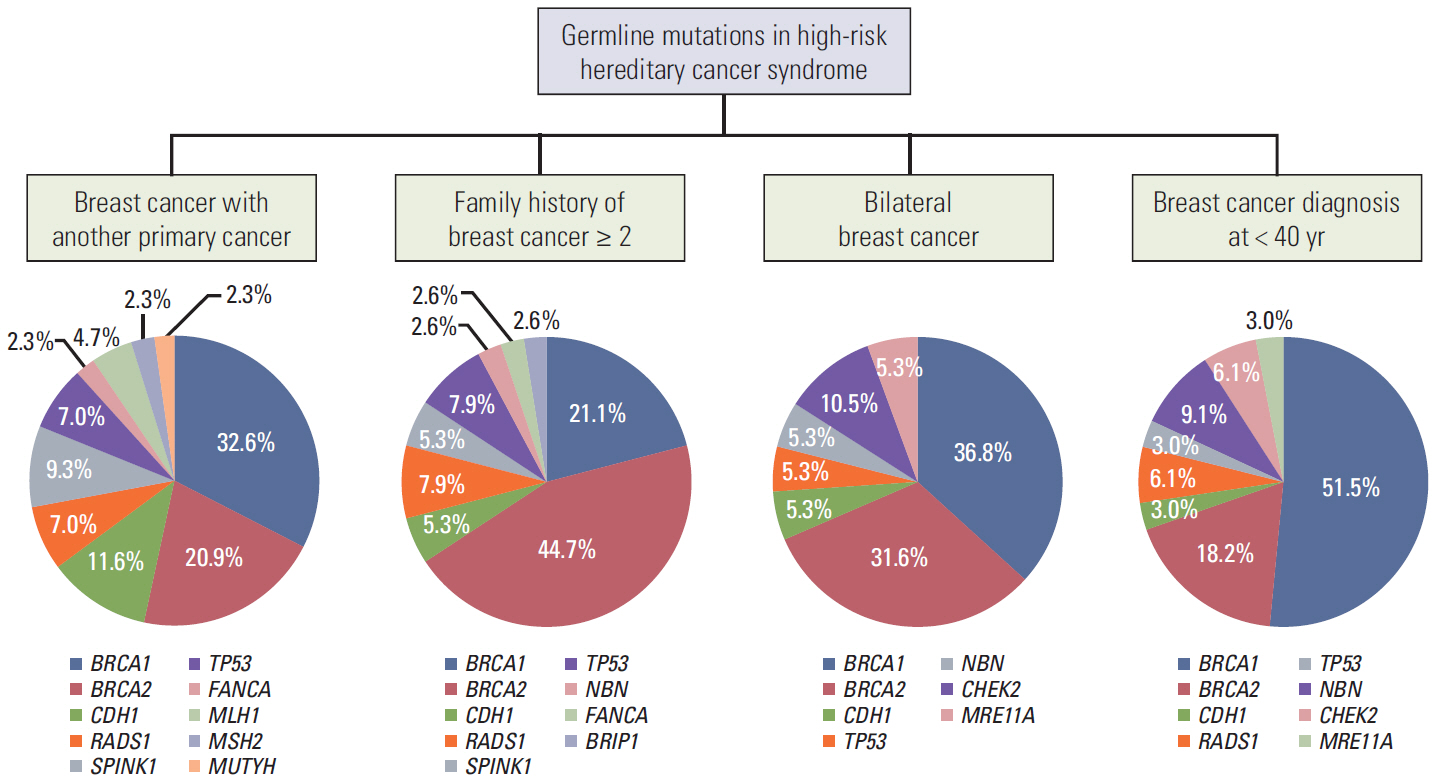
Of 496 patients, 95 patients (19.2%) were found to have 48 deleterious germline mutations in 16 cancer susceptibility genes. The deleterious mutations were found in 39 of 250 patients (15.6%) who had breast cancer and another primary cancer, 38 of 169 patients (22.5%) who had a family history of breast cancer (≥ 2 relatives), 16 of 57 patients (28.1%) who had bilateral breast cancer, and 29 of 84 patients (34.5%) who were diagnosed with breast cancer at younger than 40 years of age. Of the 95 patients with deleterious mutations, 60 patients (63.2%) had BRCA1/2 mutations and 38 patients (40.0%) had non-BRCA1/2 mutations. We detected two novel deleterious mutations in BRCA2 and MLH1.
Conclusion
NGS-based multiple-gene panel testing improved the detection rates of deleterious mutations and provided a cost-effective cancer risk assessment.
Keyword
Figure
Reference
-
References
1. Kuchenbaecker KB, Hopper JL, Barnes DR, Phillips KA, Mooij TM, Roos-Blom MJ, et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA. 2017; 317:2402–16.2. Mai PL, Khincha PP, Loud JT, DeCastro RM, Bremer RC, Peters JA, et al. Prevalence of cancer at baseline screening in the national cancer institute Li-Fraumeni syndrome cohort. JAMA Oncol. 2017; 3:1640–5.
Article3. Ricker C, Culver JO, Lowstuter K, Sturgeon D, Sturgeon JD, Chanock CR, et al. Increased yield of actionable mutations using multi-gene panels to assess hereditary cancer susceptibility in an ethnically diverse clinical cohort. Cancer Genet. 2016; 209:130–7.
Article4. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015; 17:405–24.
Article5. Hall MJ, Reid JE, Burbidge LA, Pruss D, Deffenbaugh AM, Frye C, et al. BRCA1 and BRCA2 mutations in women of different ethnicities undergoing testing for hereditary breast-ovarian cancer. Cancer. 2009; 115:2222–33.6. Kim H, Cho DY, Choi DH, Oh M, Shin I, Park W, et al. Frequency of pathogenic germline mutation in CHEK2, PALB2, MRE11, and RAD50 in patients at high risk for hereditary breast cancer. Breast Cancer Res Treat. 2017; 161:95–102.
Article7. Thompson ER, Rowley SM, Li N, McInerny S, Devereux L, Wong-Brown MW, et al. Panel testing for familial breast cancer: calibrating the tension between research and clinical care. J Clin Oncol. 2016; 34:1455–9.
Article8. Li J, Meeks H, Feng BJ, Healey S, Thorne H, Makunin I, et al. Targeted massively parallel sequencing of a panel of putative breast cancer susceptibility genes in a large cohort of multiple-case breast and ovarian cancer families. J Med Genet. 2016; 53:34–42.
Article9. Fitzgerald RC, Hardwick R, Huntsman D, Carneiro F, Guilford P, Blair V, et al. Hereditary diffuse gastric cancer: updated consensus guidelines for clinical management and directions for future research. J Med Genet. 2010; 47:436–44.
Article10. Masciari S, Larsson N, Senz J, Boyd N, Kaurah P, Kandel MJ, et al. Germline E-cadherin mutations in familial lobular breast cancer. J Med Genet. 2007; 44:726–31.
Article11. Cousineau I, Abaji C, Belmaaza A. BRCA1 regulates RAD51 function in response to DNA damage and suppresses spontaneous sister chromatid replication slippage: implications for sister chromatid cohesion, genome stability, and carcinogenesis. Cancer Res. 2005; 65:11384–91.
Article12. Depienne C, Bouteiller D, Meneret A, Billot S, Groppa S, Klebe S, et al. RAD51 haploinsufficiency causes congenital mirror movements in humans. Am J Hum Genet. 2012; 90:301–7.
Article13. Martin RW, Orelli BJ, Yamazoe M, Minn AJ, Takeda S, Bishop DK. RAD51 up-regulation bypasses BRCA1 function and is a common feature of BRCA1-deficient breast tumors. Cancer Res. 2007; 67:9658–65.
Article14. Maacke H, Jost K, Opitz S, Miska S, Yuan Y, Hasselbach L, et al. DNA repair and recombination factor Rad51 is over-expressed in human pancreatic adenocarcinoma. Oncogene. 2000; 19:2791–5.
Article15. Buisson R, Dion-Cote AM, Coulombe Y, Launay H, Cai H, Stasiak AZ, et al. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat Struct Mol Biol. 2010; 17:1247–54.
Article16. Witt H, Luck W, Hennies HC, Classen M, Kage A, Lass U, et al. Mutations in the gene encoding the serine protease inhibitor, Kazal type 1 are associated with chronic pancreatitis. Nat Genet. 2000; 25:213–6.
Article17. Midha S, Sreenivas V, Kabra M, Chattopadhyay TK, Joshi YK, Garg PK. Genetically determined chronic pancreatitis but not alcoholic pancreatitis is a strong risk factor for pancreatic cancer. Pancreas. 2016; 45:1478–84.
Article18. Zhen DB, Rabe KG, Gallinger S, Syngal S, Schwartz AG, Goggins MG, et al. BRCA1, BRCA2, PALB2, and CDKN2A mutations in familial pancreatic cancer: a PACGENE study. Genet Med. 2015; 17:569–77.
Article19. Villani A, Shore A, Wasserman JD, Stephens D, Kim RH, Druker H, et al. Biochemical and imaging surveillance in germline TP53 mutation carriers with Li-Fraumeni syndrome: 11 year follow-up of a prospective observational study. Lancet Oncol. 2016; 17:1295–305.
Article20. Seong MW, Kim KH, Chung IY, Kang E, Lee JW, Park SK, et al. A multi-institutional study on the association between BRCA1/BRCA2 mutational status and triple-negative breast cancer in familial breast cancer patients. Breast Cancer Res Treat. 2014; 146:63–9.
Article21. Krammer J, Pinker-Domenig K, Robson ME, Gonen M, Bernard-Davila B, Morris EA, et al. Breast cancer detection and tumor characteristics in BRCA1 and BRCA2 mutation carriers. Breast Cancer Res Treat. 2017; 163:565–71.
Article22. Maxwell KN, Wubbenhorst B, D'Andrea K, Garman B, Long JM, Powers J, et al. Prevalence of mutations in a panel of breast cancer susceptibility genes in BRCA1/2-negative patients with early-onset breast cancer. Genet Med. 2015; 17:630–8.
Article23. Li JY, Jing R, Wei H, Wang M, Xiaowei Q, Liu H, et al. Germline mutations in 40 cancer susceptibility genes among Chinese patients with high hereditary risk breast cancer. Int J Cancer. 2019; 144:281–9.
Article24. Antoniou AC, Casadei S, Heikkinen T, Barrowdale D, Pylkas K, Roberts J, et al. Breast-cancer risk in families with mutations in PALB2. N Engl J Med. 2014; 371:497–506.
Article25. Li N, McInerny S, Zethoven M, Cheasley D, Lim BW, Rowley SM, et al. Combined tumor sequencing and case-control analyses of RAD51C in breast cancer. J Natl Cancer Inst. 2019; 111:1332–8.
Article26. Carreira A, Kowalczykowski SC. Two classes of BRC repeats in BRCA2 promote RAD51 nucleoprotein filament function by distinct mechanisms. Proc Natl Acad Sci U S A. 2011; 108:10448–53.
Article27. Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006; 126:297–308.28. Findlay GM, Daza RM, Martin B, Zhang MD, Leith AP, Gasperini M, et al. Accurate classification of BRCA1 variants with saturation genome editing. Nature. 2018; 562:217–22.
Article29. Stadler ZK, Schrader KA, Vijai J, Robson ME, Offit K. Cancer genomics and inherited risk. J Clin Oncol. 2014; 32:687–98.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Clinical Validity of Next-Generation Sequencing Multi-Gene Panel Testing for Detecting Pathogenic Variants in Patients With Hereditary Breast-Ovarian Cancer Syndrome
- Detection of Germline Mutations in Patients with Epithelial Ovarian Cancer Using Multi-gene Panels: Beyond BRCA1/2
- Utility of Next-Generation Sequencing Panel Including Hereditary Breast and Ovarian Cancer-Related Genes for Pathogenic Variant Detection
- Diverse genetic spectrum among patients who met the criteria of hereditary breast, ovarian and pancreatic cancer syndrome
- Hereditary cancer and genetic counseling