Korean J Gastroenterol.
2020 May;75(5):231-239. 10.4166/kjg.2020.75.5.231.
Gut Microbiota and Pancreatobiliary System
- Affiliations
-
- 1Division of Gastroenterology and Hepatology, Department of Internal Medicine, Konyang University College of Medicine, Daejeon, Korea
- KMID: 2504243
- DOI: http://doi.org/10.4166/kjg.2020.75.5.231
Abstract
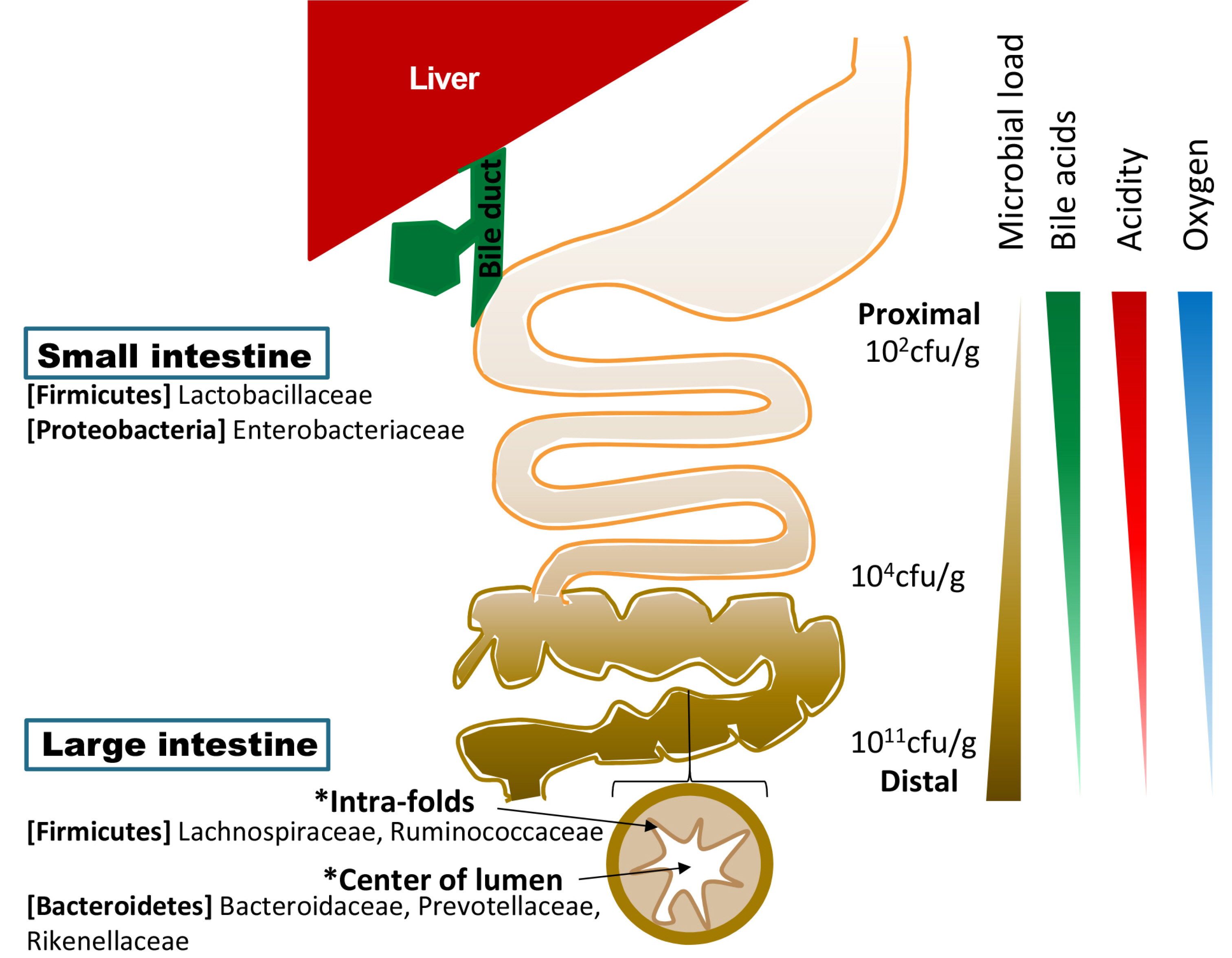
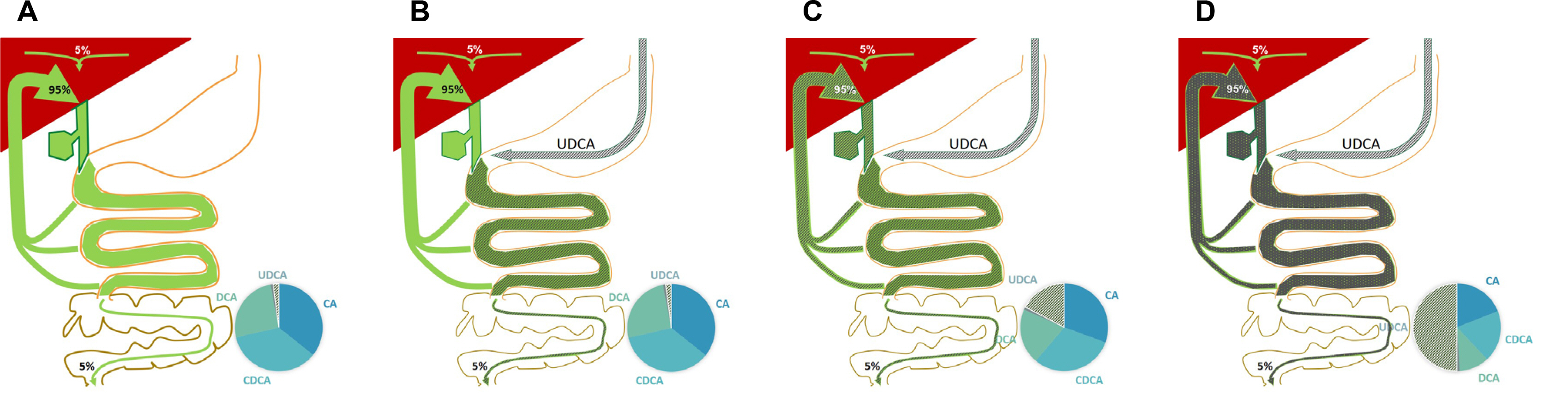
- The gut microbiota is part of the human body that is involved in body metabolism and the occurrence of various diseases. Detecting and analyzing their genetic information (microbiome) is as important as analyzing human genes. The core microbiome, the key functional genes shared by all humans, helps better understand the physiology of the human body. Information on the gut microbiome of a diseased person can help diagnose and treat disease. The pancreatobiliary system releases functional antimicrobial substances, such as bile acids and antimicrobial peptides, which affect the gut microbiota directly. In response, the gut microbiota influences pancreatobiliary secretion by controlling the generation and emission of substances through indirect signaling. This crosstalk maintains homeostasis of the pancreatobiliary system secretion and microbiota. Dysbiosis and disease can occur if this fails to work properly. Bile acid therapy has been used widely and may affect the microbial environment in the intestine. An association of the gut microbiota has been reported in many cases of pancreatobiliary diseases, including malignant tumors. Traditionally, most pancreatobiliary diseases are accompanied by infections from the gut microbiota, which is an important target for treatment. The pancreatobiliary system can control its function through physical and drug therapy. This may be a new pioneering field in the study or treatment of the gut microbiota.
Keyword
Figure
Reference
-
1. Gilbert JA, Dupont CL. 2011; Microbial metagenomics: beyond the genome. Ann Rev Mar Sci. 3:347–371. DOI: 10.1146/annurev-marine-120709-142811. PMID: 21329209.
Article2. Relman DA, Falkow S. 2001; The meaning and impact of the human genome sequence for microbiology. Trends Microbiol. 9:206–208. DOI: 10.1016/S0966-842X(01)02041-8. PMID: 11336835.
Article3. Grice EA, Segre JA. 2012; The human microbiome: our second genome. Annu Rev Genomics Hum Genet. 13:151–170. DOI: 10.1146/annurev-genom-090711-163814. PMID: 22703178. PMCID: PMC3518434.
Article4. Human Microbiome Project Consortium. 2012; Structure, function and diversity of the healthy human microbiome. Nature. 486:207–214. DOI: 10.1038/nature11234. PMID: 22699609. PMCID: PMC3564958.5. Jiménez E, Sánchez B, Farina A, Margolles A, Rodríguez JM. 2014; Characterization of the bile and gall bladder microbiota of healthy pigs. Microbiologyopen. 3:937–949. DOI: 10.1002/mbo3.218. PMID: 25336405. PMCID: PMC4263516.
Article6. Shen H, Ye F, Xie L, et al. 2015; Metagenomic sequencing of bile from gallstone patients to identify different microbial community patterns and novel biliary bacteria. Sci Rep. 5:17450. DOI: 10.1038/srep17450. PMID: 26625708. PMCID: PMC4667190.
Article7. Bravo-Blas A, Utriainen L, Clay SL, et al. 2019; Salmonella enterica serovar typhimurium travels to mesenteric lymph nodes both with host cells and autonomously. J Immunol. 202:260–267. DOI: 10.4049/jimmunol.1701254. PMID: 30487173. PMCID: PMC6305795.
Article8. Hart PA, Zen Y, Chari ST. 2015; Recent advances in autoimmune pancreatitis. Gastroenterology. 149:39–51. DOI: 10.1053/j.gastro.2015.03.010. PMID: 25770706.
Article9. Chen B, Fu SW, Lu L, Zhao H. 2019; A preliminary study of biliary microbiota in patients with bile duct stones or distal cholangiocarcinoma. Biomed Res Int. 2019:1092563. DOI: 10.1155/2019/1092563. PMID: 31662965. PMCID: PMC6778921.
Article10. Gutiérrez-Díaz I, Molinero N, Cabrera A, et al. 2018; Diet: cause or consequence of the microbial profile of cholelithiasis disease? Nutrients. 10:E1307. DOI: 10.3390/nu10091307. PMID: 30223526. PMCID: PMC6163750.
Article11. Wu T, Zhang Z, Liu B, et al. 2013; Gut microbiota dysbiosis and bacterial community assembly associated with cholesterol gallstones in large-scale study. BMC Genomics. 14:669. DOI: 10.1186/1471-2164-14-669. PMID: 24083370. PMCID: PMC3851472.
Article12. Ye F, Shen H, Li Z, et al. 2016; Influence of the biliary system on biliary bacteria revealed by bacterial communities of the human biliary and upper digestive tracts. PLoS One. 11:e0150519. DOI: 10.1371/journal.pone.0150519. PMID: 26930491. PMCID: PMC4773253.
Article13. Avilés-Jiménez F, Guitron A, Segura-López F, et al. 2016; Microbiota studies in the bile duct strongly suggest a role for Helicobacter pylori in extrahepatic cholangiocarcinoma. Clin Microbiol Infect. 22:178.e11–178.e22. DOI: 10.1016/j.cmi.2015.10.008. PMID: 26493848.
Article14. Kim SC, Tonkonogy SL, Albright CA, et al. 2005; Variable phenotypes of enterocolitis in interleukin 10-deficient mice monoassociated with two different commensal bacteria. Gastroenterology. 128:891–906. DOI: 10.1053/j.gastro.2005.02.009. PMID: 15825073.
Article15. Marteau P, Seksik P, Shanahan F. 2003; Manipulation of the bacterial flora in inflammatory bowel disease. Best Pract Res Clin Gastroenterol. 17:47–61. DOI: 10.1053/bega.2002.0344. PMID: 12617882.
Article16. Lepage P, Leclerc MC, Joossens M, et al. 2013; A metagenomic insight into our gut's microbiome. Gut. 62:146–158. DOI: 10.1136/gutjnl-2011-301805. PMID: 22525886.
Article17. Turnbaugh PJ, Hamady M, Yatsunenko T, et al. 2009; A core gut microbiome in obese and lean twins. Nature. 457:480–484. DOI: 10.1038/nature07540. PMID: 19043404. PMCID: PMC2677729.
Article18. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. 2006; An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 444:1027–1031. DOI: 10.1038/nature05414. PMID: 17183312.
Article19. Lakhan SE, Kirchgessner A. 2010; Gut inflammation in chronic fatigue syndrome. Nutr Metab (Lond). 7:79. DOI: 10.1186/1743-7075-7-79. PMID: 20939923. PMCID: PMC2964729.
Article20. Larsen N, Vogensen FK, van den Berg FW, et al. 2010; Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One. 5:e9085. DOI: 10.1371/journal.pone.0009085. PMID: 20140211. PMCID: PMC2816710.
Article21. Tilg H, Moschen AR. 2014; Microbiota and diabetes: an evolving relationship. Gut. 63:1513–1521. DOI: 10.1136/gutjnl-2014-306928. PMID: 24833634.
Article22. Castellarin M, Warren RL, Freeman JD, et al. 2012; Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 22:299–306. DOI: 10.1101/gr.126516.111. PMID: 22009989. PMCID: PMC3266037.
Article23. Kostic AD, Gevers D, Pedamallu CS, et al. 2012; Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 22:292–298. DOI: 10.1101/gr.126573.111. PMID: 22009990. PMCID: PMC3266036.
Article24. Buffington SA, Di Prisco GV, Auchtung TA, Ajami NJ, Petrosino JF, Costa-Mattioli M. 2016; Microbial reconstitution reverses maternal dietinduced social and synaptic deficits in offspring. Cell. 165:1762–1775. DOI: 10.1016/j.cell.2016.06.001. PMID: 27315483. PMCID: PMC5102250.
Article25. Kang DW, Adams JB, Coleman DM, et al. 2019; Long-term benefit of microbiota transfer therapy on autism symptoms and gut microbiota. Sci Rep. 9:5821. DOI: 10.1038/s41598-019-42183-0. PMID: 30967657. PMCID: PMC6456593.
Article26. Sampson TR, Debelius JW, Thron T, et al. 2016; Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson's disease. Cell. 167:1469–1480.e12. DOI: 10.1016/j.cell.2016.11.018. PMID: 27912057. PMCID: PMC5718049.
Article27. Huse SM, Ye Y, Zhou Y, Fodor AA. 2012; A core human microbiome as viewed through 16S rRNA sequence clusters. PLoS One. 7:e34242. DOI: 10.1371/journal.pone.0034242. PMID: 22719824. PMCID: PMC3374614.
Article28. Shetty SA, Hugenholtz F, Lahti L, Smidt H, de Vos WM. 2017; Intestinal microbiome landscaping: insight in community assemblage and implications for microbial modulation strategies. FEMS Microbiol Rev. 41:182–199. DOI: 10.1093/femsre/fuw045. PMID: 28364729. PMCID: PMC5399919.
Article29. Dethlefsen L, McFall-Ngai M, Relman DA. 2007; An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 449:811–818. DOI: 10.1038/nature06245. PMID: 17943117.
Article30. Donaldson GP, Lee SM, Mazmanian SK. 2016; Gut biogeography of the bacterial microbiota. Nat Rev Microbiol. 14:20–32. DOI: 10.1038/nrmicro3552. PMID: 26499895. PMCID: PMC4837114.
Article31. Sasatomi K, Noguchi K, Sakisaka S, Sata M, Tanikawa K. 1998; Abnormal accumulation of endotoxin in biliary epithelial cells in primary biliary cirrhosis and primary sclerosing cholangitis. J Hepatol. 29:409–416. DOI: 10.1016/S0168-8278(98)80058-5. PMID: 9764987.
Article32. Hopf U, Möller B, Stemerowicz R, et al. 1989; Relation between Escherichia coli R(rough)-forms in gut, lipid A in liver, and primary biliary cirrhosis. Lancet. 2:1419–1422. DOI: 10.1016/S0140-6736(89)92034-5. PMID: 2574361.
Article33. van Velkinburgh JC, Gunn JS. 1999; PhoP-PhoQ-regulated loci are required for enhanced bile resistance in Salmonella spp. Infect Immun. 67:1614–1622. DOI: 10.1128/IAI.67.4.1614-1622.1999. PMID: 10084994. PMCID: PMC96504.
Article34. Leverrier P, Dimova D, Pichereau V, Auffray Y, Boyaval P, Jan G. 2003; Susceptibility and adaptive response to bile salts in Propionibacterium freudenreichii: physiological and proteomic analysis. Appl Environ Microbiol. 69:3809–3818. DOI: 10.1128/AEM.69.7.3809-3818.2003. PMID: 12839748. PMCID: PMC165135.
Article35. Noh DO, Gilliland SE. 1993; Influence of bile on cellular integrity and beta-galactosidase activity of Lactobacillus acidophilus. J Dairy Sci. 76:1253–1259. DOI: 10.3168/jds.S0022-0302(93)77454-8. PMID: 8505417.36. Schubert R, Jaroni H, Schoelmerich J, Schmidt KH. 1983; Studies on the mechanism of bile salt-induced liposomal membrane damage. Digestion. 28:181–190. DOI: 10.1159/000198984. PMID: 6365666.
Article37. Fernández Murga ML, Bernik D, Font de Valdez G, Disalvo AE. 1999; Permeability and stability properties of membranes formed by lipids extracted from Lactobacillus acidophilus grown at different temperatures. Arch Biochem Biophys. 364:115–121. DOI: 10.1006/abbi.1998.1093. PMID: 10087172.38. Kandell RL, Bernstein C. 1991; Bile salt/acid induction of DNA damage in bacterial and mammalian cells: implications for colon cancer. Nutr Cancer. 16:227–238. DOI: 10.1080/01635589109514161. PMID: 1775385.
Article39. García-Quintanilla M, Ramos-Morales F, Casadesús J. 2008; Conjugal transfer of the Salmonella enterica virulence plasmid in the mouse intestine. J Bacteriol. 190:1922–1927. DOI: 10.1128/JB.01626-07. PMID: 18178735. PMCID: PMC2258861.40. Prieto AI, Ramos-Morales F, Casadesús J. 2004; Bile-induced DNA damage in Salmonella enterica. Genetics. 168:1787–1794. DOI: 10.1534/genetics.104.031062. PMID: 15611156. PMCID: PMC1448704.
Article41. Flahaut S, Frere J, Boutibonnes P, Auffray Y. 1996; Comparison of the bile salts and sodium dodecyl sulfate stress responses in Enterococcus faecalis. Appl Environ Microbiol. 62:2416–2420. DOI: 10.1128/AEM.62.7.2416-2420.1996. PMID: 8779581. PMCID: PMC168024.
Article42. Rajagopalan N, Lindenbaum S. 1982; The binding of Ca2+ to taurine and glycine-conjugated bile salt micelles. Biochim Biophys Acta. 711:66–74. DOI: 10.1016/0005-2760(82)90010-8. PMID: 7066374.
Article43. Sanyal AJ, Hirsch JI, Moore EW. 1994; Premicellar taurocholate enhances calcium uptake from all regions of rat small intestine. Gastroenterology. 106:866–874. DOI: 10.1016/0016-5085(94)90744-7. PMID: 8143992.
Article44. Dominguez DC. 2004; Calcium signalling in bacteria. Mol Microbiol. 54:291–297. DOI: 10.1111/j.1365-2958.2004.04276.x. PMID: 15469503.
Article45. Maloney PR, Parks DJ, Haffner CD, et al. 2000; Identification of a chemical tool for the orphan nuclear receptor FXR. J Med Chem. 43:2971–2974. DOI: 10.1021/jm0002127. PMID: 10956205.
Article46. Inagaki T, Moschetta A, Lee YK, et al. 2006; Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci U S A. 103:3920–3925. DOI: 10.1073/pnas.0509592103. PMID: 16473946. PMCID: PMC1450165.
Article47. Bajaj JS, Heuman DM, Hylemon PB, et al. 2014; Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol. 60:940–947. DOI: 10.1016/j.jhep.2013.12.019. PMID: 24374295. PMCID: PMC3995845.
Article48. Begley M, Gahan CG, Hill C. 2005; The interaction between bacteria and bile. FEMS Microbiol Rev. 29:625–651. DOI: 10.1016/j.femsre.2004.09.003. PMID: 16102595.
Article49. Vavassori P, Mencarelli A, Renga B, Distrutti E, Fiorucci S. 2009; The bile acid receptor FXR is a modulator of intestinal innate immunity. J Immunol. 183:6251–6261. DOI: 10.4049/jimmunol.0803978. PMID: 19864602.
Article50. Matsubara T, Li F, Gonzalez FJ. 2013; FXR signaling in the enterohepatic system. Mol Cell Endocrinol. 368:17–29. DOI: 10.1016/j.mce.2012.05.004. PMID: 22609541. PMCID: PMC3491147.
Article51. Li T, Chiang JY. 2014; Bile acid signaling in metabolic disease and drug therapy. Pharmacol Rev. 66:948–983. DOI: 10.1124/pr.113.008201. PMID: 25073467. PMCID: PMC4180336.
Article52. Makishima M, Okamoto AY, Repa JJ, et al. 1999; Identification of a nuclear receptor for bile acids. Science. 284:1362–1365. DOI: 10.1126/science.284.5418.1362. PMID: 10334992.
Article53. D'Aldebert E, Biyeyeme Bi Mve MJ, Mergey M, et al. 2009; Bile salts control the antimicrobial peptide cathelicidin through nuclear receptors in the human biliary epithelium. Gastroenterology. 136:1435–1443. DOI: 10.1053/j.gastro.2008.12.040. PMID: 19245866.54. Gombart AF. 2009; The vitamin D-antimicrobial peptide pathway and its role in protection against infection. Future Microbiol. 4:1151–1165. DOI: 10.2217/fmb.09.87. PMID: 19895218. PMCID: PMC2821804.
Article55. Hofmann AF, Eckmann L. 2006; How bile acids confer gut mucosal protection against bacteria. Proc Natl Acad Sci U S A. 103:4333–4334. DOI: 10.1073/pnas.0600780103. PMID: 16537368. PMCID: PMC1450168.
Article56. Northfield TC, McColl I. 1973; Postprandial concentrations of free and conjugated bile acids down the length of the normal human small intestine. Gut. 14:513–518. DOI: 10.1136/gut.14.7.513. PMID: 4729918. PMCID: PMC1412809.
Article57. Bansal S, Singh M, Kidwai S, et al. 2014; Bile acid amphiphiles with tunable head groups as highly selective antitubercular agents. MedChemComm. 5:1761–1768. DOI: 10.1039/C4MD00303A.
Article58. Qi Y, Jiang C, Cheng J, et al. 2015; Bile acid signaling in lipid metabolism:metabolomic and lipidomic analysis of lipid and bile acid markers linked to anti-obesity and anti-diabetes in mice. Biochim Biophys Acta. 1851:19–29. DOI: 10.1016/j.bbalip.2014.04.008. PMID: 24796972. PMCID: PMC4219936.59. Nevens F, Andreone P, Mazzella G, et al. 2016; A placebo-controlled trial of obeticholic acid in primary biliary cholangitis. N Engl J Med. 375:631–643. DOI: 10.1056/NEJMoa1509840. PMID: 27532829.60. Bevins CL, Salzman NH. 2011; Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. 9:356–368. DOI: 10.1038/nrmicro2546. PMID: 21423246.
Article61. Sun J, Furio L, Mecheri R, et al. 2015; Pancreatic β-cells limit autoimmune diabetes via an immunoregulatory antimicrobial peptide expressed under the influence of the gut microbiota. Immunity. 43:304–317. DOI: 10.1016/j.immuni.2015.07.013. PMID: 26253786.
Article62. Ahuja M, Schwartz DM, Tandon M, et al. 2017; Orai1-mediated antimicrobial secretion from pancreatic acini shapes the gut microbiome and regulates gut innate immunity. Cell Metab. 25:635–646. DOI: 10.1016/j.cmet.2017.02.007. PMID: 28273482. PMCID: PMC5345693.
Article63. Stenwall A, Ingvast S, Skog O, Korsgren O. 2019; Characterization of host defense molecules in the human pancreas. Islets. 11:89–101. DOI: 10.1080/19382014.2019.1585165. PMID: 31242128. PMCID: PMC6682263.
Article64. Hogan PG. 2015; The STIM1-ORAI1 microdomain. Cell Calcium. 58:357–367. DOI: 10.1016/j.ceca.2015.07.001. PMID: 26215475. PMCID: PMC4564343.
Article65. Kakiyama G, Pandak WM, Gillevet PM, et al. 2013; Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol. 58:949–955. DOI: 10.1016/j.jhep.2013.01.003. PMID: 23333527. PMCID: PMC3936319.
Article66. Kim DB, Paik CN, Lee JM, Kim YJ. 2020; Association between increased breath hydrogen methane concentration and prevalence of glucose intolerance in acute pancreatitis. J Breath Res. 14:026006. DOI: 10.1088/1752-7163/ab5460. PMID: 31689699.
Article67. Kim DB, Paik CN, Sung HJ, et al. 2015; Breath hydrogen and methane are associated with intestinal symptoms in patients with chronic pancreatitis. Pancreatology. 15:514–518. DOI: 10.1016/j.pan.2015.07.005. PMID: 26278025.
Article68. Kim DB, Paik CN, Song DS, Kim YJ, Lee JM. 2018; The characteristics of small intestinal bacterial overgrowth in patients with gallstone diseases. J Gastroenterol Hepatol. 33:1477–1484. DOI: 10.1111/jgh.14113. PMID: 29392773.
Article69. Sung HJ, Paik CN, Chung WC, Lee KM, Yang JM, Choi MG. 2015; Small intestinal bacterial overgrowth diagnosed by glucose hydrogen breath test in post-cholecystectomy patients. J Neurogastroenterol Motil. 21:545–551. DOI: 10.5056/jnm15020. PMID: 26351251. PMCID: PMC4622137.
Article70. Nakamoto N, Sasaki N, Aoki R, et al. 2019; Gut pathobionts underlie intestinal barrier dysfunction and liver T helper 17 cell immune response in primary sclerosing cholangitis. Nat Microbiol. 4:492–503. DOI: 10.1038/s41564-018-0333-1. PMID: 30643240.
Article71. Piddock LJ. 2006; Multidrug-resistance efflux pumps - not just for resistance. Nat Rev Microbiol. 4:629–636. DOI: 10.1038/nrmicro1464. PMID: 16845433.
Article72. Lin J, Sahin O, Michel LO, Zhang Q. 2003; Critical role of multidrug efflux pump CmeABC in bile resistance and in vivo colonization of Campylobacter jejuni. Infect Immun. 71:4250–4259. DOI: 10.1128/IAI.71.8.4250-4259.2003. PMID: 12874300. PMCID: PMC165992.
Article73. Kullak-Ublick GA, Stieger B, Meier PJ. 2004; Enterohepatic bile salt transporters in normal physiology and liver disease. Gastroenterology. 126:322–342. DOI: 10.1053/j.gastro.2003.06.005. PMID: 14699511.
Article74. Glavinas H, Krajcsi P, Cserepes J, Sarkadi B. 2004; The role of ABC transporters in drug resistance, metabolism and toxicity. Curr Drug Deliv. 1:27–42. DOI: 10.2174/1567201043480036. PMID: 16305368.
Article75. Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR. 2008; Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci U S A. 105:13580–13585. DOI: 10.1073/pnas.0804437105. PMID: 18757757. PMCID: PMC2533232.
Article76. Geller LT, Barzily-Rokni M, Danino T, et al. 2017; Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 357:1156–1160. DOI: 10.1126/science.aah5043. PMID: 28912244. PMCID: PMC5727343.
Article77. Thomas RM, Gharaibeh RZ, Gauthier J, et al. 2018; Intestinal microbiota enhances pancreatic carcinogenesis in preclinical models. Carcinogenesis. 39:1068–1078. DOI: 10.1093/carcin/bgy073. PMID: 29846515. PMCID: PMC6067127.
Article78. Pushalkar S, Hundeyin M, Daley D, et al. 2018; The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. 8:403–416. DOI: 10.1158/2159-8290.CD-17-1134. PMID: 29567829. PMCID: PMC6225783.79. Diehl GE, Longman RS, Zhang JX, et al. 2013; Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature. 494:116–120. DOI: 10.1038/nature11809. PMID: 23334413. PMCID: PMC3711636.80. Widdison AL, Karanjia ND, Reber HA. 1994; Routes of spread of pathogens into the pancreas in a feline model of acute pancreatitis. Gut. 35:1306–1310. DOI: 10.1136/gut.35.9.1306. PMID: 7959243. PMCID: PMC1375713.
Article81. Ammori BJ, Leeder PC, King RF, et al. 1999; Early increase in intestinal permeability in patients with severe acute pancreatitis: correlation with endotoxemia, organ failure, and mortality. J Gastrointest Surg. 3:252–262. DOI: 10.1016/S1091-255X(99)80067-5. PMID: 10481118.
Article82. Sonika U, Goswami P, Thakur B, et al. 2017; Mechanism of increased intestinal permeability in acute pancreatitis: alteration in tight junction proteins. J Clin Gastroenterol. 51:461–466. DOI: 10.1097/MCG.0000000000000612. PMID: 27466164.83. Liu H, Li W, Wang X, Li J, Yu W. 2008; Early gut mucosal dysfunction in patients with acute pancreatitis. Pancreas. 36:192–196. DOI: 10.1097/MPA.0b013e31815a399f. PMID: 18376312.
Article84. Madhani K, Farrell JJ. 2016; Autoimmune pancreatitis: an update on diagnosis and management. Gastroenterol Clin North Am. 45:29–43. DOI: 10.1016/j.gtc.2015.10.005. PMID: 26895679.85. Hamada S, Masamune A, Nabeshima T, Shimosegawa T. 2018; Differences in gut microbiota profiles between autoimmune pancreatitis and chronic pancreatitis. Tohoku J Exp Med. 244:113–117. DOI: 10.1620/tjem.244.113. PMID: 29434076.
Article86. de Goffau MC, Fuentes S, van den Bogert B, et al. 2014; Aberrant gut microbiota composition at the onset of type 1 diabetes in young children. Diabetologia. 57:1569–1577. DOI: 10.1007/s00125-014-3274-0. PMID: 24930037.
Article87. de Goffau MC, Luopajärvi K, Knip M, et al. 2013; Fecal microbiota composition differs between children with β-cell autoimmunity and those without. Diabetes. 62:1238–1244. DOI: 10.2337/db12-0526. PMID: 23274889. PMCID: PMC3609581.
Article88. Vatanen T, Franzosa EA, Schwager R, et al. 2018; The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature. 562:589–594. DOI: 10.1038/s41586-018-0620-2. PMID: 30356183. PMCID: PMC6296767.
Article89. Ren Z, Jiang J, Xie H, et al. 2017; Gut microbial profile analysis by MiSeq sequencing of pancreatic carcinoma patients in China. Oncotarget. 8:95176–95191. DOI: 10.18632/oncotarget.18820. PMID: 29221120. PMCID: PMC5707014.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Revolutionizing gut health: exploring the role of gut microbiota and the potential of microbiome-based therapies in lower gastrointestinal diseases
- Microbial Modulation in Inflammatory Bowel Diseases
- Alteration of Gut Microbiota in Autism Spectrum Disorder: An Overview
- Gut Microbiome and Colorectal Cancer
- Current Status and Prospects of Intestinal Microbiome Studies