Blood Res.
2020 Jun;55(2):107-111. 10.5045/br.2020.2020042.
Impact of using genotyping to predict SERF negative phenotype in Thai blood donor populations
- Affiliations
-
- 1Graduate Program in Biomedical Sciences, Faculty of Allied Health Sciences, Thammasat University, Pathumtani, Thailand
- 2Blood Bank Section, Faculty of Medicine, Chiang Mai University, Chiang Mai, Thailand
- 3Regional Blood Centre 12th Songkhla, Thai Red Cross Society, Songkhla, Thailand
- 4National Blood Centre, Thai Red Cross Society, Bangkok, Thailand
- KMID: 2503485
- DOI: http://doi.org/10.5045/br.2020.2020042
Abstract
- Background
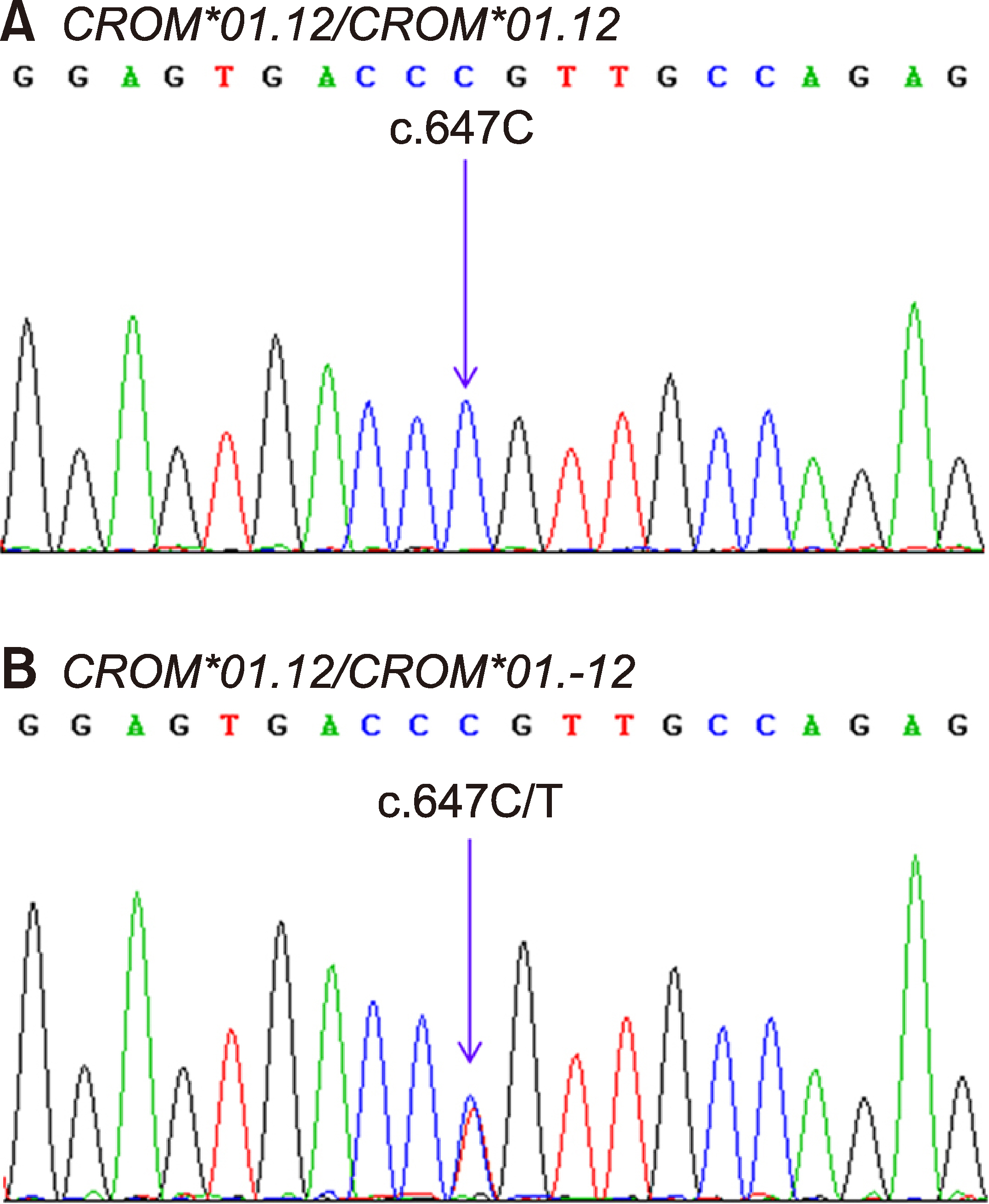
SERF(+) is a high prevalence antigen in the Cromer blood group system that is encoded by a CROM*01.12 allele. The SERF(-) on red cells is caused by a single nucleotide variation, c.647C>T, in exon 5 of the Decay-accelerating factor, DAF gene. Alloanti-SERF was found in a pregnant Thai woman, and a SERF(-) individual was found among Thai blood donors. Since anti-SERF is commercially unavailable, this study aimed to develop appropriate genotyping methods for CROM*01.12 and CROM*01.-12 alleles and predict the SERF(-) phenotype in Thai blood donors.
Methods
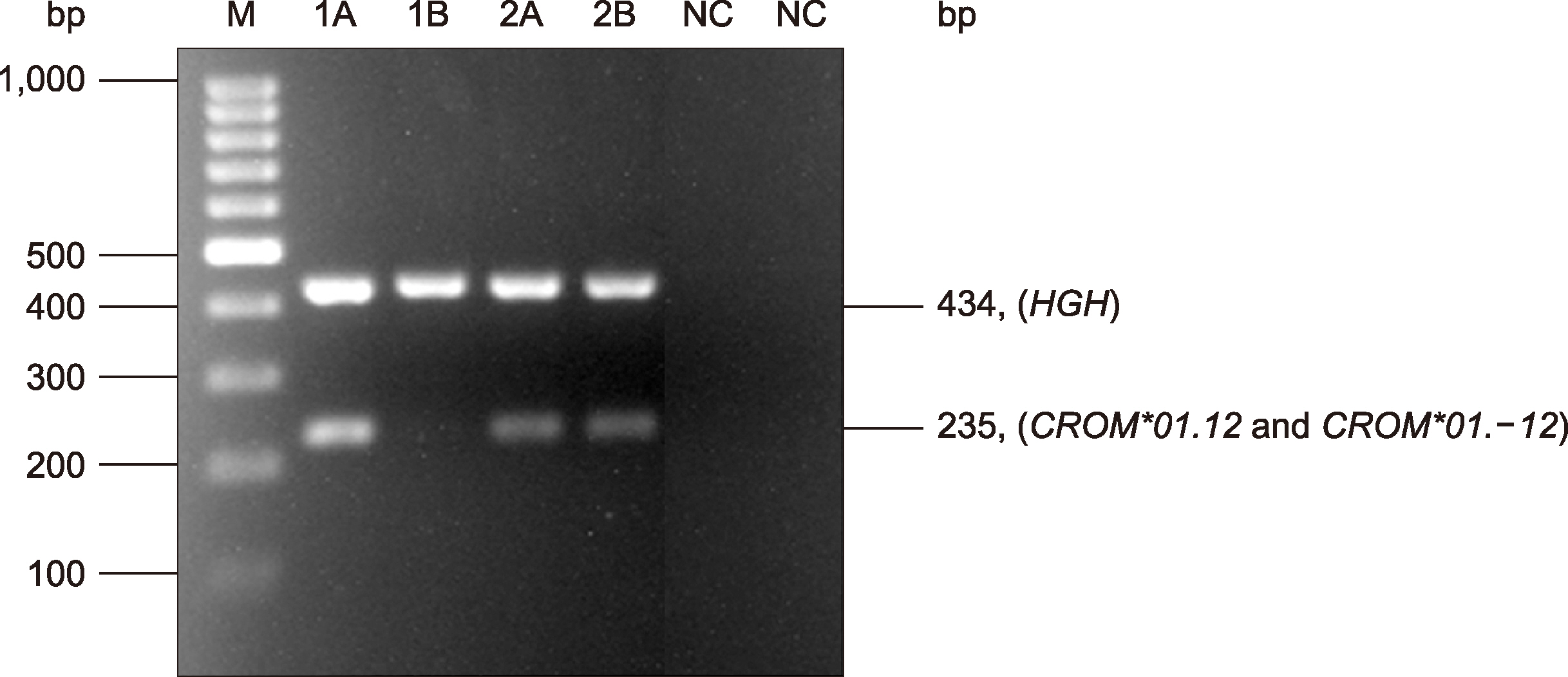
DNA samples obtained from 1,580 central, 300 northern, and 427 southern Thai blood donors were genotyped for CROM*01.12 and CROM*01.-12 allele detection using in-house PCR with sequence-specific primer (PCR-SSP) confirmed by DNA sequencing.
Results
Validity of the PCR-SSP genotyping results agreed with DNA sequencing; CROM*01.12/ CROM*01.12 was the most common (98.42%, 98.00%, and 98.59%), followed by CROM*01.12/CROM*01.-12 (1.58%, 2.00%, and 1.41%) among central, northern, and southern Thais, respectively. CROM*01.-12/CROM*01.-12 was not detected in all three populations. The alleles found in central Thais did not significantly differ from those found in northern and southern Thais.
Conclusion
This study is the first to distinguish the predicted SERF phenotypes from genotyping results obtained using in-house PCR-SSP, confirming that the CROM*01.-12 allele frequency ranged from 0.007 to 0.010 in three Thai populations. This helps identify the SERF(-) phenotype among donors and patients, ultimately preventing adverse transfusion reactions.
Figure
Reference
-
1. Storry JR, Clausen FB, Castilho L, et al. 2019; International Society of Blood Transfusion Working Party on Red Cell Immunogenetics and Blood Group Terminology: Report of the Dubai, Copenhagen and Toronto meetings. Vox Sang. 114:95–102. DOI: 10.1111/vox.12717. PMID: 30421425. PMCID: PMC6342652.
Article2. Banks J, Poole J, Ahrens N, et al. 2004; SERF: a new antigen in the Cromer blood group system. Transfus Med. 14:313–8. DOI: 10.1111/j.0958-7578.2004.00519.x. PMID: 15285728.
Article3. Storry JR, Reid ME, Yazer MH. 2010; The Cromer blood group system: a review. Immunohematology. 26:109–18. PMID: 21214297.
Article4. Palacajornsuk P, Hue-Roye K, Nathalang O, Tantimavanich S, Bejrachandra S, Reid ME. 2005; Analysis of SERF in Thai blood donors. Immunohematology. 21:66–9. PMID: 15954807.
Article5. Kupatawintu P, Emthip M, Sungnoon D, et al. 2010; Unexpected antibodies of patients, blood samples sent for testing at NBC. TRCS. J Hematol Transfus Med. 20:255–62.6. Deelert S, Thippayaboon P, Sriwai W, et al. 2010; Jk(a-b-) phenotype screening by the urea lysis test in Thai blood donors. Blood Transfus. 8:17–20.7. Nathalang O, Intharanut K, Sasikarn W, Nathalang S, Kupatawintu P. 2016; A new polymerase chain reaction: sequence-specific primer method for the Augustine blood type. Blood Transfus. 14:577–9.8. Fung MK, Eder AF, Spitalnik SL, Westhoff CM, editors. 2017. Technical manual. 19th ed. AABB;Bethesda, MD:9. Avent ND. 2009; Large-scale blood group genotyping: clinical implications. Br J Haematol. 144:3–13. DOI: 10.1111/j.1365-2141.2008.07285.x. PMID: 19016734.10. Wagner FF, Bittner R, Petershofen EK, Doescher A, Müller TH. 2008; Cost-efficient sequence-specific priming-polymerase chain reaction screening for blood donors with rare phenotypes. Transfusion. 48:1169–73. DOI: 10.1111/j.1537-2995.2008.01682.x. PMID: 18422854.
Article11. Gassner C, Meyer S, Frey BM, Vollmert C. 2013; Matrix-assisted laser desorption/ionisation, time-of-flight mass spectrometry-based blood group genotyping--the alternative approach. Transfus Med Rev. 27:2–9. DOI: 10.1016/j.tmrv.2012.10.001. PMID: 23217607.
Article12. Latini FR, Gazito D, Arnoni CP, et al. 2014; A new strategy to identify rare blood donors: single polymerase chain reaction multiplex SNaPshot reaction for detection of 16 blood group alleles. Blood Transfus. 12(Suppl 1):s256–63. DOI: 10.2450/2013.0242-12. PMID: 23736910. PMCID: PMC3934258.13. Hong YJ, Chung Y, Hwang SM, et al. 2016; Genotyping of 22 blood group antigen polymorphisms and establishing a national recipient registry in the Korean population. Ann Hematol. 95:985–91. DOI: 10.1007/s00277-016-2645-7. PMID: 27021300.
Article14. Liu Z, Zeng R, Chen Q, et al. 2012; Genotyping for Kidd, Kell, Duffy, Scianna, and RHCE blood group antigens polymorphisms in Jiangsu Chinese Han. Chin Med J (Engl). 125:1076–81. PMID: 22613534.15. Reid M, Lomas-Francis C, Olsson M, editors. 2012. The blood group antigen factsbook. 3rd ed. Academic Press;London, UK: DOI: 10.1016/B978-0-12-415849-8.00001-6.