Clin Endosc.
2020 Mar;53(2):127-131. 10.5946/ce.2020.046.
Lesion-Based Convolutional Neural Network in Diagnosis of Early Gastric Cancer
- Affiliations
-
- 1Division of Gastroenterology, Department of Internal Medicine, Soonchunhyang University College of Medicine, Cheonan, Korea
- 2Division of Gastroenterology, Department of Internal Medicine, Gangnam Severance Hospital, Yonsei University College of Medicine, Seoul, Korea
- KMID: 2500881
- DOI: http://doi.org/10.5946/ce.2020.046
Abstract
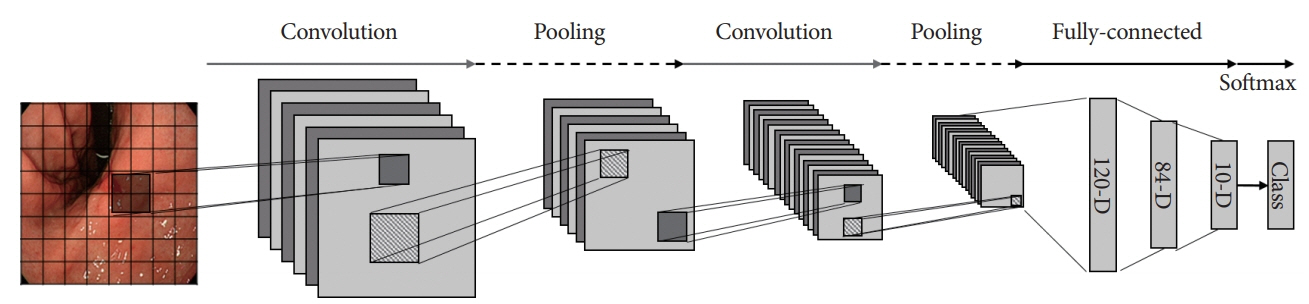
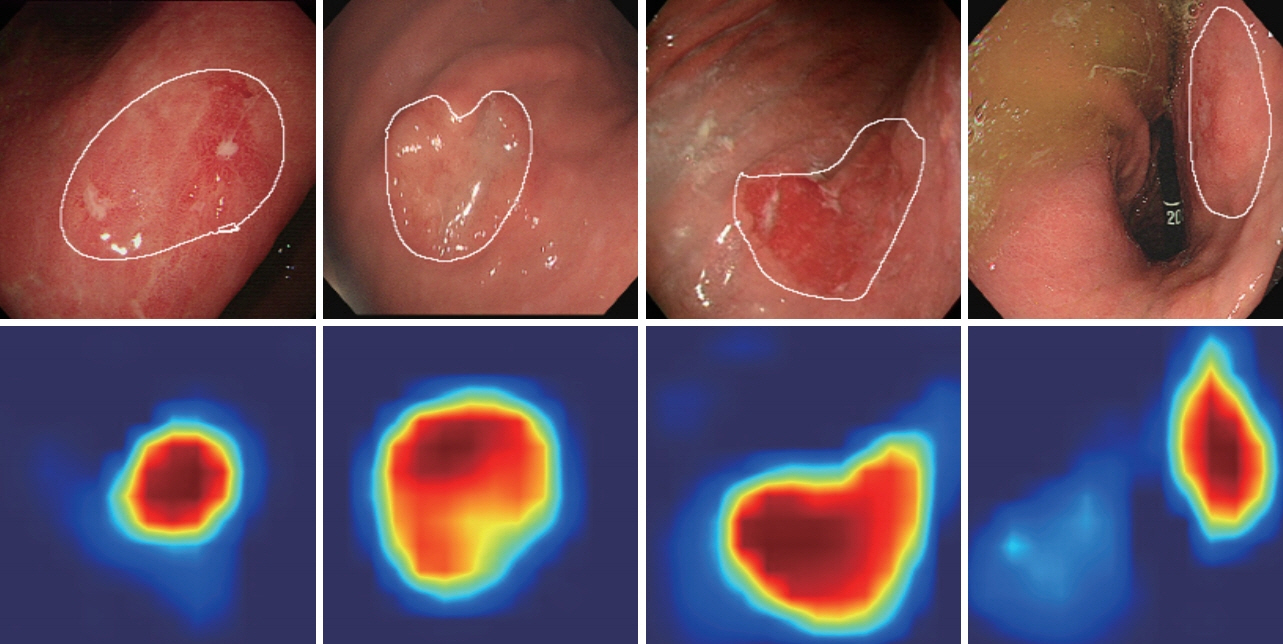
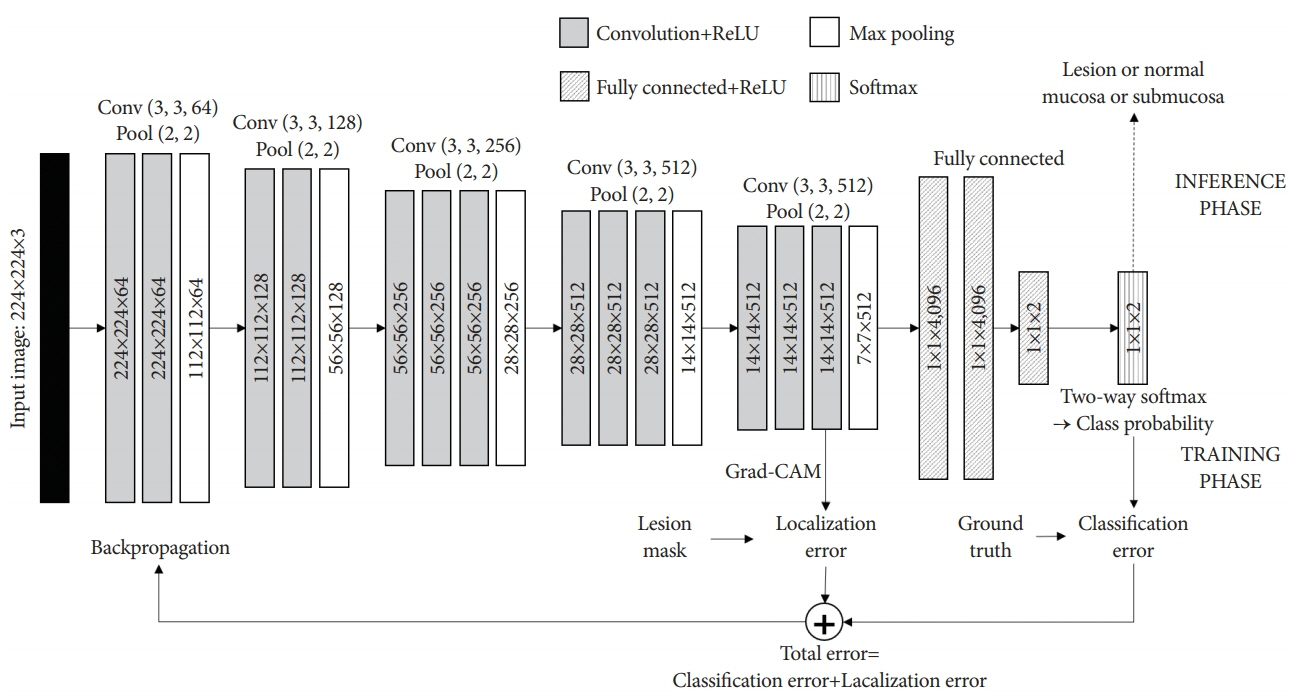
- Diagnosis and evaluation of early gastric cancer (EGC) using endoscopic images is significantly important; however, it has some limitations. In several studies, the application of convolutional neural network (CNN) greatly enhanced the effectiveness of endoscopy. To maximize clinical usefulness, it is important to determine the optimal method of applying CNN for each organ and disease. Lesion�-based CNN is a type of deep learning model designed to learn the entire lesion from endoscopic images. This review describes the application of lesion-based CNN technology in diagnosis of EGC.
Keyword
Figure
Reference
-
1. Take I, Shi Q, Zhong Y-S. Progress with each passing day: role of endoscopy in early gastric cancer. Transl Gastrointest Cancer. 2015; 4:423–428.2. Zhu R, Zhang R, Xue D. Lesion detection of endoscopy images based on convolutional neural network features. In : In: 2015 8th International Congress on Image and Signal Processing (CISP); 2015 Oct 14-16; Shenyang, China. Piscataway (NJ). IEEE. 2015. p. 372–376.
Article3. Du W, Rao N, Liu D, et al. Review on the applications of deep learning in the analysis of gastrointestinal endoscopy images. IEEE Access. 2019; 7:142053–142069.
Article4. Lecun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proceedings of the IEEE. 1998; 86:2278–2324.
Article5. Hirasawa T, Aoyama K, Tanimoto T, et al. Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer. 2018; 21:653–660.
Article6. Horie Y, Yoshio T, Aoyama K, et al. Diagnostic outcomes of esophageal cancer by artificial intelligence using convolutional neural networks. Gastrointest Endosc. 2019; 89:25–32.
Article7. Urban G, Tripathi P, Alkayali T, et al. Deep learning localizes and identifies polyps in real time with 96% accuracy in screening colonoscopy. Gastroenterology. 2018; 155:1069–1078. e8.
Article8. Zhu Y, Wang QC, Xu MD, et al. Application of convolutional neural network in the diagnosis of the invasion depth of gastric cancer based on conventional endoscopy. Gastrointest Endosc. 2019; 89:806–815. e1.
Article9. Krizhevsky A, Sutskever I, Hinton GE, et al. Imagenet classification with deep convolutional neural networks. Advances in Neural Information Processing Systems. 2012; 1097–1105.
Article10. Fukushima K. Neocognitron: a self organizing neural network model for a mechanism of pattern recognition unaffected by shift in position. Biol Cybern. 1980; 36:193–202.11. Lo SB, Lou SA, Lin JS, Freedman MT, Chien MV, Mun SK. Artificial convolution neural network techniques and applications for lung nodule detection. IEEE Trans Med Imaging. 1995; 14:711–718.
Article12. Han SS, Park GH, Lim W, et al. Deep neural networks show an equivalent and often superior performance to dermatologists in onychomycosis diagnosis: automatic construction of onychomycosis datasets by region-based convolutional deep neural network. PLoS One. 2018; 13:e0191493.
Article13. Larson DB, Chen MC, Lungren MP, Halabi SS, Stence NV, Langlotz CP. Performance of a deep-learning neural network model in assessing skeletal maturity on pediatric hand radiographs. Radiology. 2018; 287:313–322.
Article14. Rajpurkar P, Irvin J, Zhu K, et al. CheXNet: radiologist-level pneumonia detection on chest x-rays with deep learning [Internet]. c2017. Available from: https://arxiv.org/abs/1711.05225.15. Mannath J, Ragunath K. Role of endoscopy in early oesophageal cancer. Nat Rev Gastroenterol Hepatol. 2016; 13:720–730.
Article16. Komeda Y, Handa H, Watanabe T, et al. Computer-aided diagnosis based on convolutional neural network system for colorectal polyp classification: preliminary experience. Oncology. 2017; 93(Suppl 1):30–34.
Article17. Zhang R, Zheng Y, Mak TW, et al. Automatic detection and classification of colorectal polyps by transferring low-level CNN features from nonmedical domain. IEEE J Biomed Health Inform. 2017; 21:41–47.
Article18. Lequan Y, Hao C, Qi D, Jing Q, Pheng Ann H. Integrating online and offline three-dimensional deep learning for automated polyp detection in colonoscopy videos. IEEE J Biomed Health Inform. 2017; 21:65–75.19. Byrne MF, Chapados N, Soudan F, et al. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut. 2019; 68:94–100.
Article20. Karnes WE, Alkayali T, Mittal M, et al. Automated polyp detection using deep learning: leveling the field. Gastrointest Endosc. 2017; 85(5 Suppl):AB376–AB377.21. Shichijo S, Nomura S, Aoyama K, et al. Application of convolutional neural networks in the diagnosis of Helicobacter pylori infection based on endoscopic images. EBioMedicine. 2017; 25:106–111.
Article22. Hong J, Park B, Park H. Convolutional neural network classifier for distinguishing Barrett’s esophagus and neoplasia endomicroscopy images. In : In: 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC); 2017 Jul 11-15; Seogwipo, Korea. Piscataway (NJ). IEEE. 2017. p. 2892–2895.
Article23. Ito N, Kawahira H, Nakashima H, Uesato M, Miyauchi H, Matsubara H. Endoscopic diagnostic support system for cT1b colorectal cancer using deep learning. Oncology. 2019; 96:44–50.
Article24. Gotoda T. Macroscopic feature of “Gastritis-like cancer” with little malignant appearance in early gastric cancer. Stomach and Intestine. 1999; 34:1495–1504.25. Chiu PWY, Uedo N, Singh R, et al. An Asian consensus on standards of diagnostic upper endoscopy for neoplasia. Gut. 2019; 68:186–197.
Article26. Yao K, Doyama H, Gotoda T, et al. Diagnostic performance and limitations of magnifying narrow-band imaging in screening endoscopy of early gastric cancer: a prospective multicenter feasibility study. Gastric Cancer. 2014; 17:669–679.
Article27. Fujiwara S, Yao K, Nagahama T, et al. Can we accurately diagnose minute gastric cancers (≤5 mm)? Chromoendoscopy (CE) vs magnifying endoscopy with narrow band imaging (M-NBI). Gastric Cancer. 2015; 18:590–596.28. Yao K. The endoscopic diagnosis of early gastric cancer. Ann Gastroenterol. 2013; 26:11–22.29. Goto O, Fujishiro M, Kodashima S, Ono S, Omata M. Outcomes of endoscopic submucosal dissection for early gastric cancer with special reference to validation for curability criteria. Endoscopy. 2009; 41:118–122.
Article30. Choi J, Kim SG, Im JP, Kim JS, Jung HC, Song IS. Comparison of endoscopic ultrasonography and conventional endoscopy for prediction of depth of tumor invasion in early gastric cancer. Endoscopy. 2010; 42:705–713.
Article31. Abe S, Oda I, Shimazu T, et al. Depth-predicting score for differentiated early gastric cancer. Gastric Cancer. 2011; 14:35–40.
Article32. Luinetti O, Fiocca R, Villani L, Alberizzi P, Ranzani GN, Solcia E. Genetic pattern, histological structure, and cellular phenotype in early and advanced gastric cancers: evidence for structure-related genetic subsets and for loss of glandular structure during progression of some tumors. Hum Pathol. 1998; 29:702–709.
Article33. Yoon HJ, Kim YH, Kim JH, et al. Are new criteria for mixed histology necessary for endoscopic resection in early gastric cancer? Pathol Res Pract. 2016; 212:410–414.
Article34. Hanaoka N, Tanabe S, Mikami T, Okayasu I, Saigenji K. Mixed-histologic- type submucosal invasive gastric cancer as a risk factor for lymph node metastasis: feasibility of endoscopic submucosal dissection. Endoscopy. 2009; 41:427–432.35. Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-CAM: visual explanations from deep networks via gradient-based localization. In : In: 2017 IEEE International Conference on Computer Vision (ICCV); 2017 Oct 22-29; Venice, Italy. Piscataway (NJ). IEEE. 2017. p. 618–626.
Article36. Yoon HJ, Kim S, Kim JH, et al. A lesion-based convolutional neural network improves endoscopic detection and depth prediction of early gastric cancer. J Clin Med. 2019; 8:E1310.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Convolutional Neural Network Based Sinogram Extrapolation for Truncated CT: Preliminary Study
- Deep learning convolutional neural network algorithms for the early detection and diagnosis of dental caries on periapical radiographs: A systematic review
- Overview of Deep Learning in Gastrointestinal Endoscopy
- Automatic Prediction of Atrial Fibrillation Based on Convolutional Neural Network Using a Short-term Normal Electrocardiogram Signal
- Endoscopic diagnosis of early gastric cancer