Cancer Res Treat.
2020 Apr;52(2):634-644. 10.4143/crt.2019.207.
Clinical Impact of Somatic Variants in Homologous RecombinationRepair-Related Genes in Ovarian High-Grade Serous Carcinoma
- Affiliations
-
- 1Department of Obstetrics and Gynecology, Comprehensive Gynecologic Cancer Center
- 2Center for Cancer Precision Medicine
- 3Department of Pathology, CHA Bundang Medical Center, CHA University, Seongnam, Korea
- KMID: 2500347
- DOI: http://doi.org/10.4143/crt.2019.207
Abstract
- Purpose
In this study, we investigated the frequencies of mutations in DNA damage repair genes including BRCA1, BRCA2, homologous recombination genes and TP53 gene in ovarian highgrade serous carcinoma, alongside those of germline and somatic BRCA mutations, with the aim of improving the identification of patients suitable for treatment with poly(ADPribose) polymerase inhibitors.
Materials and Methods
Tissue samples from 77 Korean patients with ovarian high-grade serous carcinoma were subjected to next-generation sequencing. Pathogenic alterations of 38 DNA damage repair genes and TP53 gene and their relationships with patient survival were examined. Additionally, we analyzed BRCA germline variants in blood samples from 47 of the patients for comparison.
Results
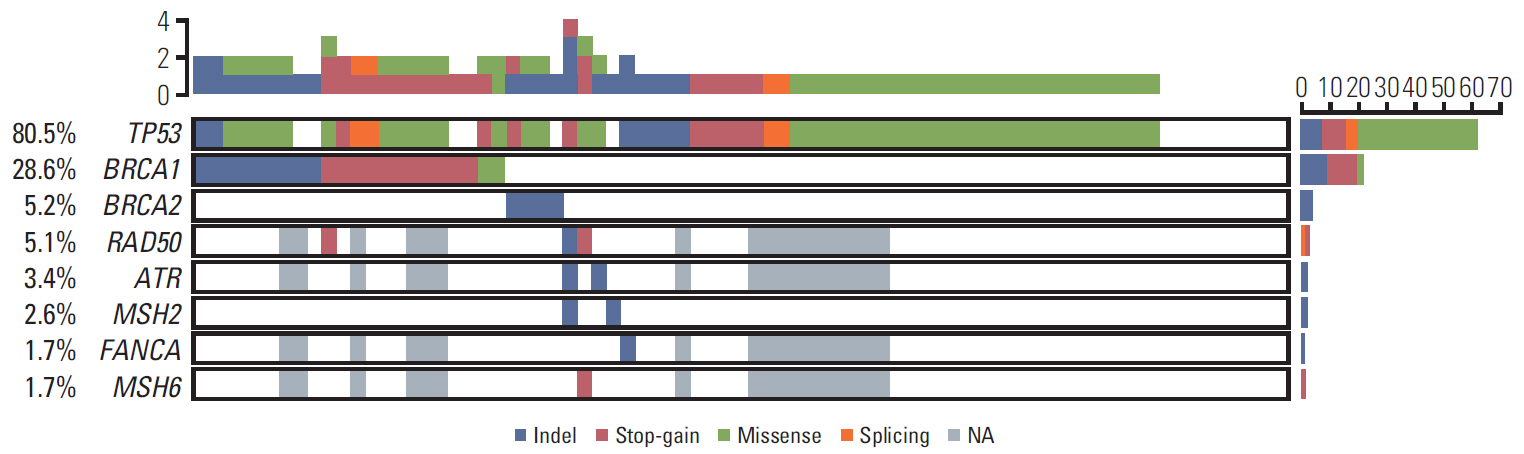
BRCA1, BRCA2, and TP53 mutations were detected in 28.6%, 5.2%, and 80.5% of the 77 patients, respectively. Alterations in RAD50, ATR, MSH6, MSH2, and FANCA were also identified. At least one mutation in a DNA damage repair gene was detected in 40.3% of patients (31/77). Germline and somatic BRCA mutations were found in 20 of 47 patients (42.6%), and four patients had only somatic mutations without germline mutations (8.5%, 4/47). Patients with DNA damage repair gene alterations with or without TP53mutation, exhibited better disease-free survival than those with TP53 mutation alone.
Conclusion
DNA damage repair genes were mutated in 40.3% of patients with high-grade serous carcinoma, with somatic BRCAmutations in the absence of germline mutation in 8.5%. Somatic variant examination, along with germline testing of DNA damage repair genes, has potential to detect additional candidates for PARP inhibitor treatment.
Keyword
Figure
Reference
-
References
1. Kurman RJ, Carcangiu ML, Herrington CS, Young RH. WHO classification of tumours of female reproductive organs. 4th ed. Lyon: International Agency for Research on Cancer;2014.2. Walsh T, Casadei S, Lee MK, Pennil CC, Nord AS, Thornton AM, et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci U S A. 2011; 108:18032–7.
Article3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018; 68:7–30.
Article4. Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011; 474:609–15.5. Fong PC, Yap TA, Boss DS, Carden CP, Mergui-Roelvink M, Gourley C, et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J Clin Oncol. 2010; 28:2512–9.6. Ledermann J, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 2014; 15:852–61.
Article7. Loveday C, Turnbull C, Ramsay E, Hughes D, Ruark E, Frankum JR, et al. Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat Genet. 2011; 43:879–82.
Article8. McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift S, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006; 66:8109–15.
Article9. Gelmon KA, Tischkowitz M, Mackay H, Swenerton K, Robidoux A, Tonkin K, et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol. 2011; 12:852–61.
Article10. Petitjean A, Achatz MI, Borresen-Dale AL, Hainaut P, Olivier M. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene. 2007; 26:2157–65.
Article11. Knijnenburg TA, Wang L, Zimmermann MT, Chambwe N, Gao GF, Cherniack AD, et al. Genomic and molecular landscape of DNA damage repair deficiency across The Cancer Genome Atlas. Cell Rep. 2018; 23:239–54.12. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010; 38:e164.
Article13. McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, et al. The Ensembl variant effect predictor. Genome Biol. 2016; 17:122.
Article14. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015; 17:405–24.
Article15. Choi MC, Heo JH, Jang JH, Jung SG, Park H, Joo WD, et al. Germline mutations of BRCA1 and BRCA2 in Korean ovarian cancer patients: finding founder mutations. Int J Gynecol Cancer. 2015; 25:1386–91.16. Przybycin CG, Kurman RJ, Ronnett BM, Shih IM, Vang R. Are all pelvic (nonuterine) serous carcinomas of tubal origin? Am J Surg Pathol. 2010; 34:1407–16.
Article17. Pennington KP, Walsh T, Harrell MI, Lee MK, Pennil CC, Rendi MH, et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin Cancer Res. 2014; 20:764–75.
Article18. Safra T, Lai WC, Borgato L, Nicoletto MO, Berman T, Reich E, et al. BRCA mutations and outcome in epithelial ovarian cancer (EOC): experience in ethnically diverse groups. Ann Oncol. 2013; 24 Suppl 8:viii63–8.
Article19. Patel JN, Braicu I, Timms KM, Solimeno C, Tshiaba P, Reid J, et al. Characterisation of homologous recombination deficiency in paired primary and recurrent high-grade serous ovarian cancer. Br J Cancer. 2018; 119:1060–6.
Article20. Ramus SJ, Gayther SA. The contribution of BRCA1 and BRCA2 to ovarian cancer. Mol Oncol. 2009; 3:138–50.21. Foster KA, Harrington P, Kerr J, Russell P, DiCioccio RA, Scott IV, et al. Somatic and germline mutations of the BRCA2 gene in sporadic ovarian cancer. Cancer Res. 1996; 56:3622–5.22. Berchuck A, Heron KA, Carney ME, Lancaster JM, Fraser EG, Vinson VL, et al. Frequency of germline and somatic BRCA1 mutations in ovarian cancer. Clin Cancer Res. 1998; 4:2433–7.23. Mafficini A, Simbolo M, Parisi A, Rusev B, Luchini C, Cataldo I, et al. BRCA somatic and germline mutation detection in paraffin embedded ovarian cancers by next-generation sequencing. Oncotarget. 2016; 7:1076–83.
Article24. Hennessy BT, Timms KM, Carey MS, Gutin A, Meyer LA, Flake DD 2nd, et al. Somatic mutations in BRCA1 and BRCA2 could expand the number of patients that benefit from poly (ADP ribose) polymerase inhibitors in ovarian cancer. J Clin Oncol. 2010; 28:3570–6.
Article25. Zhao Q, Yang J, Li L, Cao D, Yu M, Shen K, et al. Germline and somatic mutations in homologous recombination genes among Chinese ovarian cancer patients detected using next-generation sequencing. J Gynecol Oncol. 2017; 28:e39.
Article26. McAlpine JN, Porter H, Kobel M, Nelson BH, Prentice LM, Kalloger SE, et al. BRCA1 and BRCA2 mutations correlate with TP53 abnormalities and presence of immune cell infiltrates in ovarian high-grade serous carcinoma. Mod Pathol. 2012; 25:740–50.
Article27. Chao A, Chang TC, Lapke N, Jung SM, Chi P, Chen CH, et al. Prevalence and clinical significance of BRCA1/2 germline and somatic mutations in Taiwanese patients with ovarian cancer. Oncotarget. 2016; 7:85529–41.
Article28. Tinker AV, Gelmon K. The role of PARP inhibitors in the treatment of ovarian carcinomas. Curr Pharm Des. 2012; 18:3770–4.
Article29. Kim SI, Lee M, Kim HS, Chung HH, Kim JW, Park NH, et al. Effect of BRCA mutational status on survival outcome in advanced-stage high-grade serous ovarian cancer. J Ovarian Res. 2019; 12:40.
Article30. Kim DH, Cho CH, Kwon SY, Ryoo NH, Jeon DS, Lee W, et al. BRCA1/2 mutations, including large genomic rearrangements, among unselected ovarian cancer patients in Korea. J Gynecol Oncol. 2018; 29:e90.
Article31. Choi MC, Bae JS, Jung SG, Park H, Joo WD, Song SH, et al. Prevalence of germline BRCA mutations among women with carcinoma of the peritoneum or fallopian tube. J Gynecol Oncol. 2018; 29:e43.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Differences of EDR Chemoresistance Assay and Prognosis between Recurrent Micropapillary Serous Ovarian Carcinoma and Serous Ovarian Carcinoma
- MAD2 Expression in Ovarian Carcinoma: Different Expression Patterns and Levels among Various Types of Ovarian Carcinoma and Its Prognostic Significance in High-Grade Serous Carcinoma
- Germline and somatic mutations in homologous recombination genes among Chinese ovarian cancer patients detected using next-generation sequencing
- Mutational Analysis of KRAS, BRAF, and TP53 Genes of Ovarian Serous Carcinomas in Korean Women
- Two Cases of Primary Papillary Serous Peritoneal Carcinoma