Investig Clin Urol.
2020 Mar;61(2):127-135. 10.4111/icu.2020.61.2.127.
Targeted next-generation sequencing for locally advanced prostate cancer in the Korean population
- Affiliations
-
- 1Department of Urology, Seoul National University College of Medicine, Seoul, Korea. mdrafael@snu.ac.kr
- 2Department of Urology, SMG-SNU Boramae Medical Center, Seoul, Korea.
- 3Department of Biomedical Research, Biomedical Research Institute, Seoul National University Hospital, Seoul, Korea.
- KMID: 2471068
- DOI: http://doi.org/10.4111/icu.2020.61.2.127
Abstract
- PURPOSE
This study aimed to evaluate the feasibility of pan-cancer panel analysis for locally advanced prostate cancer in the Korean population.
MATERIALS AND METHODS
We analyzed 20 patients with locally advanced prostate cancer who underwent radical prostatectomy. A pan-cancer panel (1.9 Mbp) developed by Seoul National University Hospital (SNUH), composed of 183 target genes, 23 fusion genes, and 45 drug target regions was used for this analysis. We compared the SNUH pan-cancer panel results with The Cancer Genome Atlas (TCGA) database to search for different mutations in the Korean population. Clinical data were analyzed with univariate and multivariate analysis, and p-values <0.05 were considered statistically significant. Kaplan-Meier curve and log-rank tests were performed to evaluate survival.
RESULTS
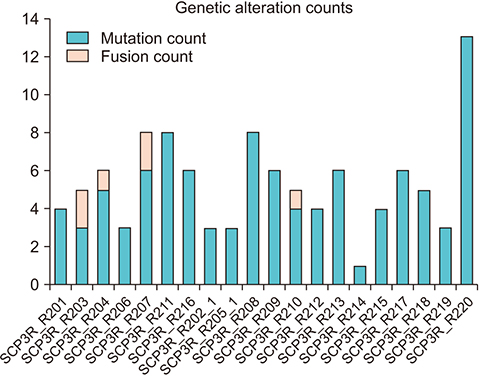
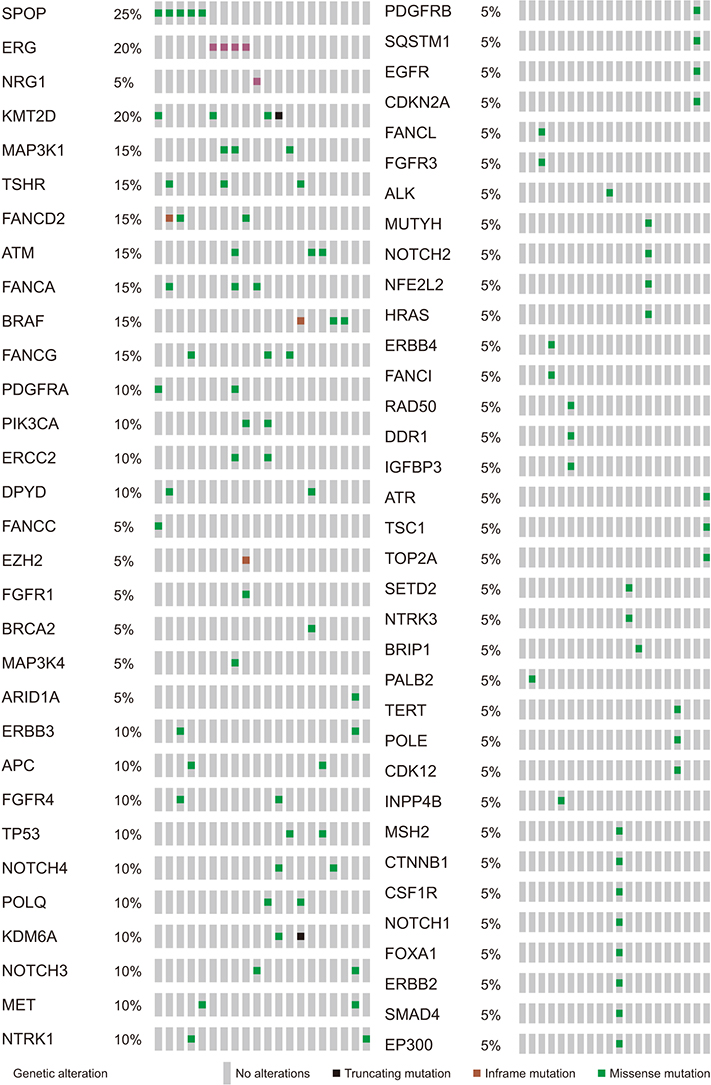
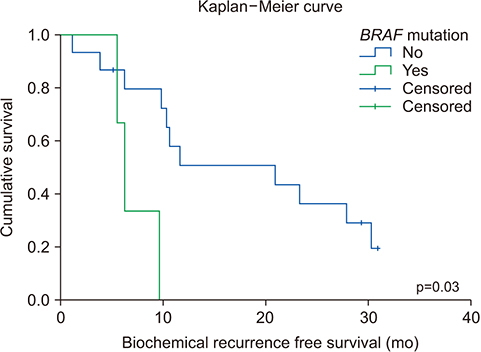
The average age of the patients and initial prostate-specific antigen values were 69.3±7.8 years and 66.3±16.9 ng/dL, respectively. Average sequencing depth was 574.5±304.1×. Ninety-nine genetic mutations and 5 fusions were detected. SPOP (25%), KMT2D (20%), and BRAF (15%) were frequently detected. ERG fusions were recurrently detected in 20% of the patients, with SLMAP and SETD4 as novel fusion partners. BRAF mutation was frequently detected in this study, but not in the TCGA database. Multivariate analysis showed BRAF mutation as an independent prognostic factor for biochemical recurrence (hazard ratio, 9.84; p=0.03).
CONCLUSIONS
The pan-cancer panel comprising genes related to prostate cancer is a useful tool for evaluating genetic alterations in locally advanced prostate cancers. Our results suggest that the BRAF mutation is associated with biochemical recurrence in the Korean population.
MeSH Terms
Figure
Reference
-
1. Roberts JS, Gornick MC, Le LQ, Bartnik NJ, Zikmund-Fisher BJ, Chinnaiyan AM. MI-ONCOSEQ Study team. Next-generation sequencing in precision oncology: patient understanding and expectations. Cancer Med. 2019; 8:227–237.
Article2. Luthra R, Patel KP, Routbort MJ, Broaddus RR, Yau J, Simien C, et al. A targeted high-throughput next-generation sequencing panel for clinical screening of mutations, gene amplifications, and fusions in solid tumors. J Mol Diagn. 2017; 19:255–264.
Article3. Kamps R, Brandão RD, Bosch BJ, Paulussen AD, Xanthoulea S, Blok MJ, et al. Next-generation sequencing in oncology: genetic diagnosis, risk prediction and cancer classification. Int J Mol Sci. 2017; 18:E308.
Article4. Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial sloan kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015; 17:251–264.5. Cheng DT, Prasad M, Chekaluk Y, Benayed R, Sadowska J, Zehir A, et al. Comprehensive detection of germline variants by MSK-IMPACT, a clinical diagnostic platform for solid tumor molecular oncology and concurrent cancer predisposition testing. BMC Med Genomics. 2017; 10:33.
Article6. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017; 67:7–30.
Article7. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015; 136:E359–E386.
Article8. Arnold M, Karim-Kos HE, Coebergh JW, Byrnes G, Antilla A, Ferlay J, et al. Recent trends in incidence of five common cancers in 26 European countries since 1988: analysis of the european cancer observatory. Eur J Cancer. 2015; 51:1164–1187.
Article9. Benafif S, Kote-Jarai Z, Eeles RA. PRACTICAL Consortium. A review of prostate cancer genome-wide association studies (GWAS). Cancer Epidemiol Biomarkers Prev. 2018; 27:845–857.
Article10. Schumacher FR, Al Olama AA, Berndt SI, Benlloch S, Ahmed M, Saunders EJ, et al. Australian Prostate Cancer BioResource (APCB). IMPACT Study. Canary PASS Investigators. Breast and Prostate Cancer Cohort Consortium (BPC3). PRACTICAL (Prostate Cancer Association Group to Investigate Cancer-Associated Alterations in the Genome) Consortium. Cancer of the Prostate in Sweden (CAPS). Prostate Cancer Genome-wide Association Study of Uncommon Susceptibility Loci (PEGASUS). Genetic Associations and Mechanisms in Oncology (GAME-ON)/Elucidating Loci Involved in Prostate Cancer Susceptibility (ELLIPSE) Consortium. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat Genet. 2018; 50:928–936.11. Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell. 2015; 163:1011–1025.12. Kumar A, Coleman I, Morrissey C, Zhang X, True LD, Gulati R, et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med. 2016; 22:369–378.
Article13. Marzec J, Mao X, Li M, Wang M, Feng N, Gou X, et al. PRACTICAL Consortium. CHIPGECS Group. A genetic study and meta-analysis of the genetic predisposition of prostate cancer in a Chinese population. Oncotarget. 2016; 7:21393–21403.
Article14. Wang M, Takahashi A, Liu F, Ye D, Ding Q, Qin C, et al. Largescale association analysis in Asians identifies new susceptibility loci for prostate cancer. Nat Commun. 2015; 6:8469.
Article15. Jeong CW, Suh J, Yuk HD, Tae BS, Kim M, Keam B, et al. Establishment of the Seoul National University Prospectively Enrolled Registry for Genitourinary Cancer (SUPER-GUC): a prospective, multidisciplinary, bio-bank linked cohort and research platform. Investig Clin Urol. 2019; 60:235–243.
Article16. Fraser M, Sabelnykova VY, Yamaguchi TN, Heisler LE, Livingstone J, Huang V, et al. Genomic hallmarks of localized, nonindolent prostate cancer. Nature. 2017; 541:359–364.17. Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015; 161:1215–1228.18. Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: a precision Oncology Knowledge Base. JCO Precis Oncol. 2017; 2017.
Article19. Carneiro A, Barbosa ÁRG, Takemura LS, Kayano PP, Moran NKS, Chen CK, et al. The role of immunohistochemical analysis as a tool for the diagnosis, prognostic evaluation and treatment of prostate cancer: a systematic review of the literature. Front Oncol. 2018; 8:377.
Article20. Lo Iacono M, Buttigliero C, Monica V, Bollito E, Garrou D, Cappia S, et al. Retrospective study testing next generation sequencing of selected cancer-associated genes in resected prostate cancer. Oncotarget. 2016; 7:14394–14404.
Article21. Ren S, Wei GH, Liu D, Wang L, Hou Y, Zhu S, et al. Whole-genome and transcriptome sequencing of prostate cancer identify new genetic alterations driving disease progression. Eur Urol. 2018; 73:322–339.
Article22. Jung KW, Won YJ, Oh CM, Kong HJ, Cho H, Lee DH, et al. Prediction of cancer incidence and mortality in Korea, 2015. Cancer Res Treat. 2015; 47:142–148.
Article23. Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002; 417:949–954.
Article24. Namba H, Nakashima M, Hayashi T, Hayashida N, Maeda S, Rogounovitch TI, et al. Clinical implication of hot spot BRAF mutation, V599E, in papillary thyroid cancers. J Clin Endocrinol Metab. 2003; 88:4393–4397.25. Liu T, Willmore-Payne C, Layfield LJ, Holden JA. Lack of BRAF activating mutations in prostate adenocarcinoma: a study of 93 cases. Appl Immunohistochem Mol Morphol. 2009; 17:121–125.26. Cho NY, Choi M, Kim BH, Cho YM, Moon KC, Kang GH. BRAF and KRAS mutations in prostatic adenocarcinoma. Int J Cancer. 2006; 119:1858–1862.27. Ren G, Liu X, Mao X, Zhang Y, Stankiewicz E, Hylands L, et al. Identification of frequent BRAF copy number gain and alterations of RAF genes in Chinese prostate cancer. Genes Chromosomes Cancer. 2012; 51:1014–1023.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Utility of Next-Generation Sequencing for Deciphering Intratumor Heterogeneity in Prostate Cancer
- Next-Generation Sequencing in Prostate Cancer
- The course forward: next generation sequencing as part of the next generation management of patients with locally advanced cervical cancer
- Rationale of Surgery in Locally Advanced and Oligometastatic Prostate Cancer
- Next generation sequencing and anti-cancer therapy