Ann Lab Med.
2020 May;40(3):224-231. 10.3343/alm.2020.40.3.224.
A Novel Heterozygous Missense Variant (c.667G>T;p.Gly223Cys) in USH1C That Interferes With Cadherin-Related 23 and Harmonin Interaction Causes Autosomal Dominant Nonsyndromic Hearing Loss
- Affiliations
-
- 1GC Genome, Yongin, Korea. changski.md@gmail.com
- 2Unité de génétique et physiologie de l'audition, Institut Pasteur, Paris, France.
- 3UMRS 1120, Inserm, Paris, France.
- 4Sorbonne Universités, Paris, France.
- 5Department of Otolaryngology - Head and Neck Surgery, Stanford University, Stanford, California, USA.
- 6College de France and Institut Pasteur, Paris, France.
- 7Department of Ophthalmology, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea.
- 8Department of Otorhinolaryngology-Head and Neck Surgery, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea.
- 9Korean Bioinformation Center, Korea Research Institute of Bioscience and Biotechnology, Daejeon, Korea.
- 10Department of Bioinformatics, University of Sciences and Technology, Daejeon, Korea.
- KMID: 2466015
- DOI: http://doi.org/10.3343/alm.2020.40.3.224
Abstract
- BACKGROUND
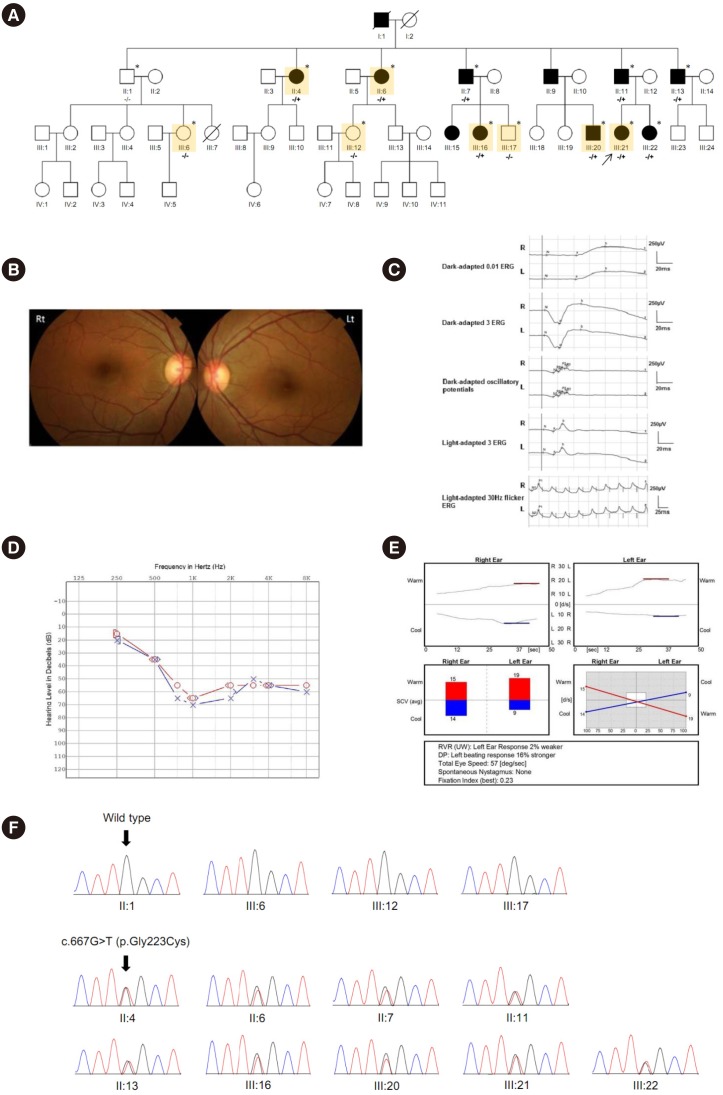
Pathogenic variants of USH1C, encoding a PDZ-domain-containing protein called harmonin, have been known to cause autosomal recessive syndromic or nonsyndromic hearing loss (NSHL). We identified a causative gene in a large Korean family with NSHL showing a typical pattern of autosomal dominant (AD) inheritance.
METHODS
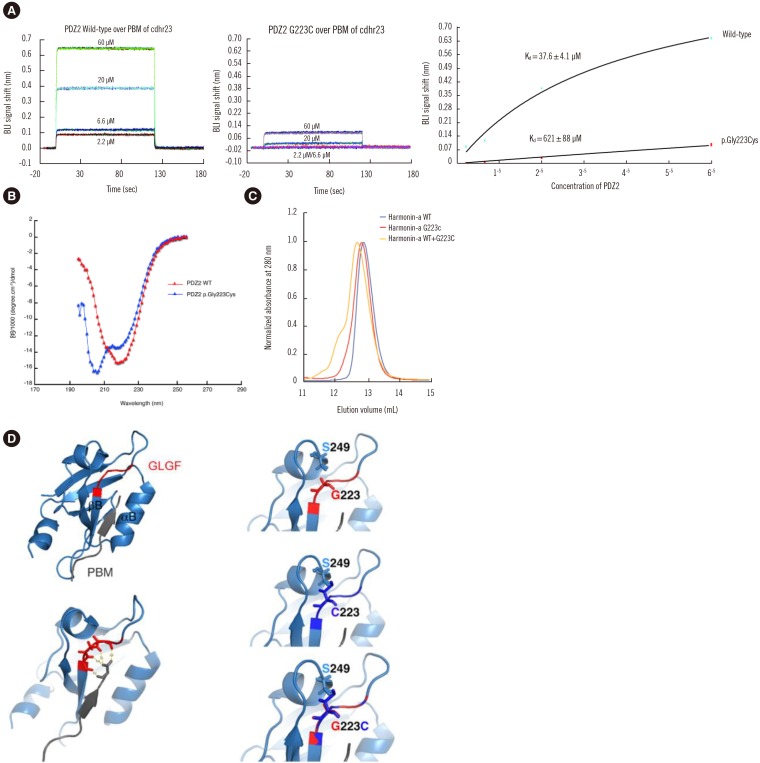
Exome sequencing was performed for five affected and three unaffected individuals in this family. Following identification of a candidate gene variant, segregation analysis and functional studies, including circular dichroism and biolayer interferometry experiments, were performed.
RESULTS
A novel USH1C heterozygous missense variant (c.667G>T;p.Gly223Cys) was shown to segregate with the NSHL phenotype in this family. This variant affects an amino acid residue located in the highly conserved carboxylate-binding loop of the harmonin PDZ2 domain and is predicted to disturb the interaction with cadherin-related 23 (cdh23). The affinity of the variant PDZ2 domain for a biotinylated synthetic peptide containing the PDZ-binding motif of cdh23 was approximately 16-fold lower than that of the wild-type PDZ2 domain and that this inaccessibility of the binding site was caused by a conformational change in the variant PDZ2 domain.
CONCLUSIONS
A heterozygous variant of USH1C that interferes with the interaction between cdh23 and harmonin causes novel AD-NSHL.
MeSH Terms
Figure
Reference
-
1. Shearer AE, Hildebrand MS, Smith RJH. Hereditary hearing loss and deafness overview. In : Adam MP, Ardinger HH, Pagon RA, editors. GeneReviews [Internet]. Seattle, WA: 1993. Updated on July 2017. https://www.ncbi.nlm.nih.gov/books/NBK1434/.2. Blaydon DC, Mueller RF, Hutchin TP, Leroy BP, Bhattacharya SS, Bird AC, et al. The contribution of USH1C mutations to syndromic and non-syndromic deafness in the UK. Clin Genet. 2003; 63:303–307. PMID: 12702164.3. Ahmed ZM, Smith TN, Riazuddin S, Makishima T, Ghosh M, Bokhari S, et al. Nonsyndromic recessive deafness DFNB18 and Usher syndrome type IC are allelic mutations of USHIC. Hum Genet. 2002; 110:527–531. PMID: 12107438.4. Ouyang XM, Xia XJ, Verpy E, Du LL, Pandya A, Petit C, et al. Mutations in the alternatively spliced exons of USH1C cause non-syndromic recessive deafness. Hum Genet. 2002; 111:26–30. PMID: 12136232.5. Khateb S, Zelinger L, Ben-Yosef T, Merin S, Crystal-Shalit O, Gross M, et al. Exome sequencing identifies a founder frameshift mutation in an alternative exon of USH1C as the cause of autosomal recessive retinitis pigmentosa with late-onset hearing loss. PLoS One. 2012; 7:e51566. PMID: 23251578.6. Saihan Z, Stabej Ple Q, Robson AG, Rangesh N, Holder GE, Moore AT, et al. Mutations in the USH1C gene associated with sector retinitis pigmentosa and hearing loss. Retina. 2011; 31:1708–1716. PMID: 21487335.7. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015; 17:405–424. PMID: 25741868.8. 1000 Genomes Project Consortium. Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015; 526:68–74. PMID: 26432245.9. Bahloul A, Pepermans E, Raynal B, Wolff N, Cordier F, England P, et al. Conformational switch of harmonin, a submembrane scaffold protein of the hair cell mechanoelectrical transduction machinery. FEBS Lett. 2017; 591:2299–2310. PMID: 28653419.10. Kamat V, Rafique A. Designing binding kinetic assay on the bio-layer interferometry (BLI) biosensor to characterize antibody-antigen interactions. Anal Biochem. 2017; 536:16–31. PMID: 28802648.11. Ahmed ZM, Frolenkov GI, Riazuddin S. Usher proteins in inner ear structure and function. Physiol Genomics. 2013; 45:987–989. PMID: 24022220.12. Wu L, Pan L, Zhang C, Zhang M. Large protein assemblies formed by multivalent interactions between cadherin23 and harmonin suggest a stable anchorage structure at the tip link of stereocilia. J Biol Chem. 2012; 287:33460–33471. PMID: 22879593.13. Cosgrove D, Zallocchi M. Usher protein functions in hair cells and photoreceptors. Int J Biochem Cell Biol. 2014; 46:80–89. PMID: 24239741.14. Reiners J, Wolfrum U. Molecular analysis of the supramolecular usher protein complex in the retina. Harmonin as the key protein of the Usher syndrome. Adv Exp Med Biol. 2006; 572:349–353. PMID: 17249595.15. Kalyoncu S, Keskin O, Gursoy A. Interaction prediction and classification of PDZ domains. BMC Bioinformatics. 2010; 11:357. PMID: 20591147.16. Siemens J, Kazmierczak P, Reynolds A, Sticker M, Littlewood-Evans A, Müller U. The Usher syndrome proteins cadherin 23 and harmonin form a complex by means of PDZ-domain interactions. Proc Natl Acad Sci U S A. 2002; 99:14946–14951. PMID: 12407180.17. Lee HJ, Zheng JJ. PDZ domains and their binding partners: structure, specificity, and modification. Cell Commun Signal. 2010; 8:8. PMID: 20509869.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Advances in Understanding the Molecular Dynamics of Autosomal Dominant Auditory Neuropathy: Unveiling a Novel DIAPH3 Gene Variant Associated With Sensorineural Hearing Loss and Bilateral Vestibular Aqueduct Enlargement
- Etiology of Hearing Loss and Genetic Hearing Loss
- A Case of Wolfram Like Disorder with Type 2 Diabetes Mellitus in an Adult
- Hyaluronan Synthase 1: A Novel Candidate Gene Associated With Late-Onset Non-syndromic Hereditary Hearing Loss
- A Case of a Family with Knuckle Pads, Deafness and Palmoplantar Hyperkeratosis