Ann Lab Med.
2020 May;40(3):209-215. 10.3343/alm.2020.40.3.209.
Evaluation of Xpert Carba-R Assay v.2 to Detect Carbapenemase Genes in Two Hospitals in Korea
- Affiliations
-
- 1Department of Laboratory Medicine and Research Institute of Bacterial Resistance, Severance Hospital, Yonsei University College of Medicine, Seoul, Korea. deyong@yuhs.ac
- 2Department of Laboratory Medicine, Gyeongsang National University Hospital, Gyeongsang National University College of Medicine, Jinju, Korea.
- 3Department of Laboratory Medicine, National Health Insurance Service Ilsan Hospital, Goyang, Korea.
- 4Department of Internal Medicine, Yonsei University College of Medicine, Seoul, Korea.
- 5Department of Internal Medicine, National Health Insurance Service Ilsan Hospital, Goyang, Korea.
- KMID: 2466013
- DOI: http://doi.org/10.3343/alm.2020.40.3.209
Abstract
- BACKGROUND
As the spread of carbapenemase-producing Enterobacteriaceae poses a critical threat to public health, rapid detection of carbapenemase genes is urgently required for prompt initiation of appropriate antimicrobial therapy and infection control. We evaluated the performance of Xpert Carba-R v.2 (Cepheid, USA) compared with that of culture-based conventional PCR.
METHODS
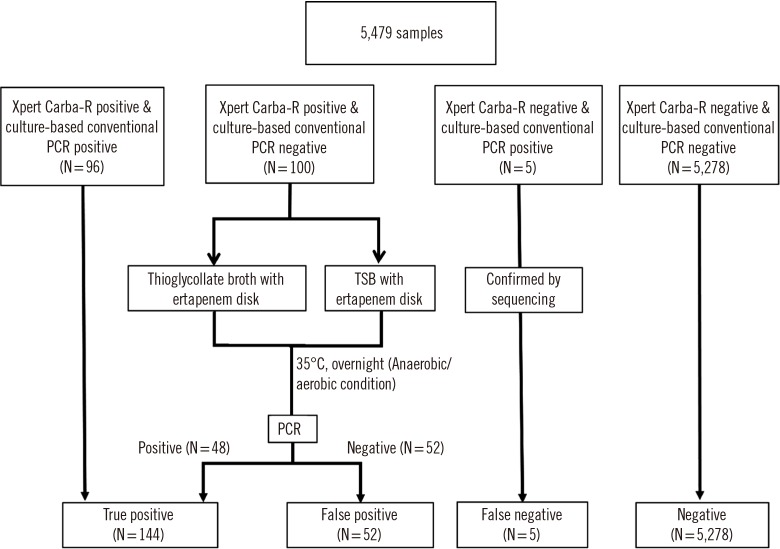
Using the results of 5,479 consecutive clinical rectal swabs, discrepant analysis (enriched culture followed by PCR) was performed for all discordant samples (N=100), which were Carba-R v.2-positive and culture-negative.
RESULTS
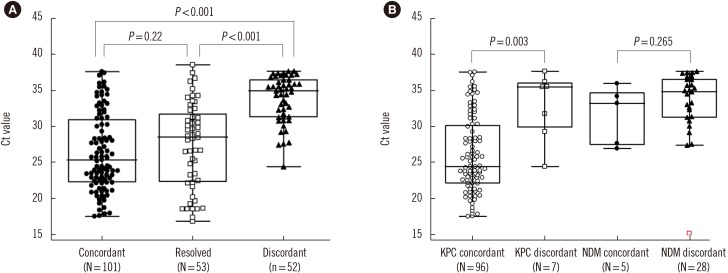
Among the samples, 206 carbapenemase genes (3.6%) were detected by Carba-R v.2. The sensitivity and specificity were 95.0% and 98.1%, respectively. The positive predictive value (PPV) and negative predictive value (NPV) were 49.0% and 99.9%, respectively. Following discrepant analysis, the PPV increased to 73.5% and the low PPV (8.1%) of the 86 non-KPC improved to 48.8%. Among the 105 discrepancies, NDM was the most frequently observed (N=56), followed by KPC (N=26), VIM (N=10), IMP (N=8), OXA-48 (N=5). The threshold cycle values between discordant vs. concordant and resolved groups were significantly different (P<0.001).
CONCLUSIONS
Carba-R v.2 is a rapid and sensitive method for detecting carbapenemase-encoding genes compared with culture-based conventional PCR. Most of our discrepant results were non-KPC genes. Thus, the clinical significance of the non-KPC positive cases detected by Carba-R v.2 should be investigated. This assay would be useful for deciding whether to isolate pre-exposed patients in hospital settings, based on the high specificity and NPV.
Keyword
MeSH Terms
Figure
Cited by 1 articles
-
국내 3차 대학병원에서 카바페넴분해효소 생성 장내세균목 검출을 위한 Xpert Carba-R Assay의 평가
Jayho Han, Sung Il Ha, Dong Pil Shin, Young Jong Cha, In Young Yoo, Yeon-Joon Park
Lab Med Online. 2022;12(3):175-182. doi: 10.47429/lmo.2022.12.3.175.
Reference
-
1. WHO. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. Geneva: World Health Organization;2017.2. Landman D, Salvani JK, Bratu S, Quale J. Evaluation of techniques for detection of carbapenem-resistant Klebsiella pneumoniae in stool surveillance cultures. J Clin Microbiol. 2005; 43:5639–5641. PMID: 16272497.3. Kim DK, Kim HS, Pinto N, Jeon J, D'Souza R, Kim MS, et al. Xpert CARBA-R assay for the detection of carbapenemase-producing organisms in intensive care unit patients of a Korean tertiary care hospital. Ann Lab Med. 2016; 36:162–165. PMID: 26709264.4. Tato M, Ruiz-Garbajosa P, Traczewski M, Dodgson A, McEwan A, Humphries R, et al. Multisite evaluation of Cepheid Xpert Carba-R assay for detection of carbapenemase-producing organisms in rectal swabs. J Clin Microbiol. 2016; 54:1814–1819. PMID: 27122379.5. Dortet L, Fusaro M, Naas T. Improvement of the Xpert Carba-R Kit for the detection of carbapenemase-producing Enterobacteriaceae. Antimicrob Agents Chemother. 2016; 60:3832–3837. PMID: 27021332.6. Uwamino Y, Sugita K, Hasegawa N, Nishimura T, Fujiwara H, Iwata S. Rapid detection and typing of carbapenemase genes from carbapenem-resistant Enterobacteriaceae isolates collected in a Japanese hospital using the Xpert Carba-R assay. Jpn J Infect Dis. 2017; 70:124–125. PMID: 27580570.7. CDC. Laboratory protocol for detection of carbapenem-resistant or carbapenemase-producing, Klebsiella spp. and E. coli from rectal swabs. Atlanta: Centers for Disease Control and Prevention;2009.8. Ahn S, Sung JY, Kim H, Kim MS, Hwang Y, Jong S, et al. Molecular epidemiology and characterization of carbapenemase-producing Enterobacteriaceae isolated at a university hospital in Korea during 4-year period. Ann Clin Microbiol. 2016; 19:39–47.9. Moore NM, Cantón R, Carretto E, Peterson LR, Sautter RL, Traczewski MM, et al. Rapid identification of five classes of carbapenem resistance genes directly from rectal swabs by use of the Xpert Carba-R assay. J Clin Microbiol. 2017; 55:2268–2275. PMID: 28515213.10. Hoyos-Mallecot Y, Ouzani S, Dortet L, Fortineau N, Naas T. Performance of the Xpert® Carba-R v2 in the daily workflow of a hygiene unit in a country with a low prevalence of carbapenemase-producing Enterobacteriaceae. Int J Antimicrob Agents. 2017; 49:774–777. PMID: 28411078.11. Schechner V, Straus-Robinson K, Schwartz D, Pfeffer I, Tarabeia J, Moskovich R, et al. Evaluation of PCR-based testing for surveillance of KPC-producing carbapenem-resistant members of the Enterobacteriaceae family. J Clin Microbiol. 2009; 47:3261–3265. PMID: 19675211.12. Schwaber MJ, Carmeli Y. An ongoing national intervention to contain the spread of carbapenem-resistant Enterobacteriaceae. Clin Infect Dis. 2014; 58:697–703. PMID: 24304707.13. Burillo A, Marín M, Cercenado E, Ruiz-Carrascoso G, Pérez-Granda MJ, Oteo J, et al. Evaluation of the Xpert Carba-R (Cepheid) assay using contrived bronchial specimens from patients with suspicion of ventilator-associated pneumonia for the detection of prevalent carbapenemases. PLoS One. 2016; 11:e0168473. PMID: 27992521.14. Otter JA, Dyakova E, Bisnauthsing KN, Querol-Rubiera A, Patel A, Ahanonu C, et al. Universal hospital admission screening for carbapenemase-producing organisms in a low-prevalence setting. J Antimicrob Chemother. 2016; 71:3556–3561. PMID: 27516471.15. Lau AF, Fahle GA, Kemp MA, Jassem AN, Dekker JP, Frank KM. Clinical Performance of Check-Direct CPE, a Multiplex PCR for direct detection of bla(KPC), bla(NDM) and/or bla(VIM), and bla(OXA)-48 from perirectal swabs. J Clin Microbiol. 2015; 53:3729–3737. PMID: 26338860.16. Tenover FC, Canton R, Kop J, Chan R, Ryan J, Weir F, et al. Detection of colonization by carbapenemase-producing Gram-negative bacilli in patients by use of the Xpert MDRO assay. J Clin Microbiol. 2013; 51:3780–3787. PMID: 24006011.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Performance of Xpert Carba-R Assay for Identification of Carbapenemase Gene in the Clinical Microbiology Laboratory
- Evaluation of the Xpert Carba-R Assay for Detection of Rectal Carbapenemase-producing Enterobacterales in a Korean Tertiary Hospital
- Xpert CARBA-R Assay for the Detection of Carbapenemase-Producing Organisms in Intensive Care Unit Patients of a Korean Tertiary Care Hospital
- Evaluation of Two Commercial Kits for Rapid Detection and Typing of Carbapenemase in Carbapenem-Resistant Enterobacterales
- Evaluation of Diagnostic Performance of RAPIDEC CARBA NP Test for Carbapenemase-Producing Enterobacteriaceae