Investig Clin Urol.
2019 Jul;60(4):227-234. 10.4111/icu.2019.60.4.227.
Genomic analysis of Korean patients with advanced prostate cancer by use of a comprehensive next-generation sequencing panel and low-coverage, whole-genome sequencing
- Affiliations
-
- 1Department of Urology, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea. hwanggyun.jeon@samsung.com
- 2Green Cross Genome, Yongin, Korea.
- 3Department of Bioinformatics, Soongsil University, Seoul, Korea.
- 4Department of Urology, Kangbuk Samsung Hospital, Sungkyunkwan University School of Medicine, Seoul, Korea.
- KMID: 2450539
- DOI: http://doi.org/10.4111/icu.2019.60.4.227
Abstract
- PURPOSE
To analyze the characteristics of somatic mutations and copy number alterations (CNAs) in Korean patients with advanced prostate cancer (PCa) by use of the Oncomine Comprehensive Panel (ThermoFisher Scientific) and low-coverage, whole-genome sequencing (LC-WGS).
MATERIALS AND METHODS
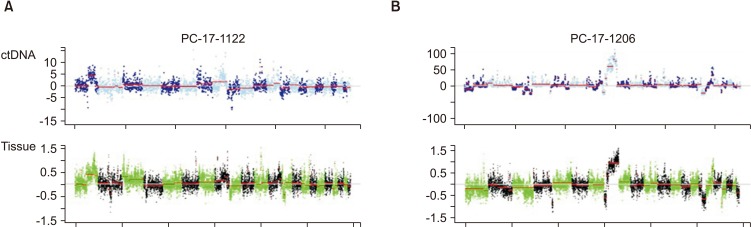
We retrospectively analyzed PCa tissues obtained from 14 patients with advanced PCa (metastatic tumor, 12 [85.7%]; nonmetastatic castration-resistant PCa, 1 [7.1%]; pT3b, 1 [7.1%]) from 2009 to 2017. The Oncomine Comprehensive Panel included a total of 143 genes. Moreover, LC-WGS was performed to detect CNAs of the entire genome. Two plasma samples matched with tumor tissues were analyzed using LC-WGS to compare the chromosomal aberration patterns between circulating tumor DNA and tumor tissue.
RESULTS
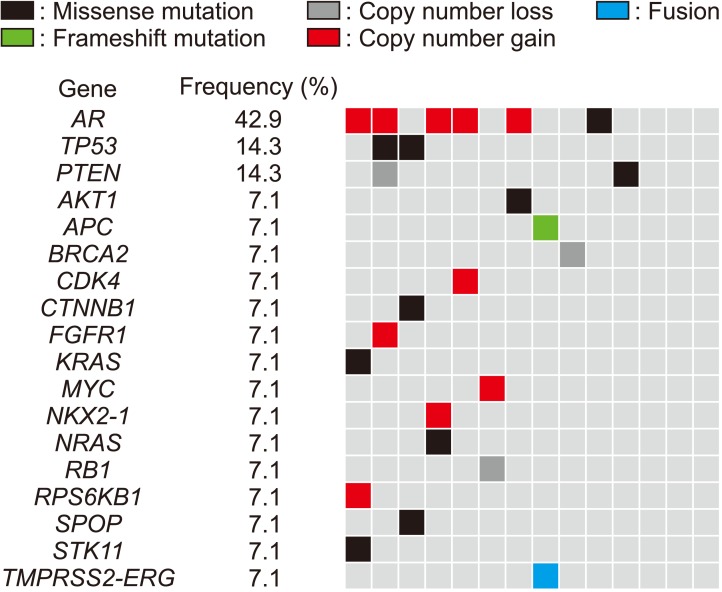
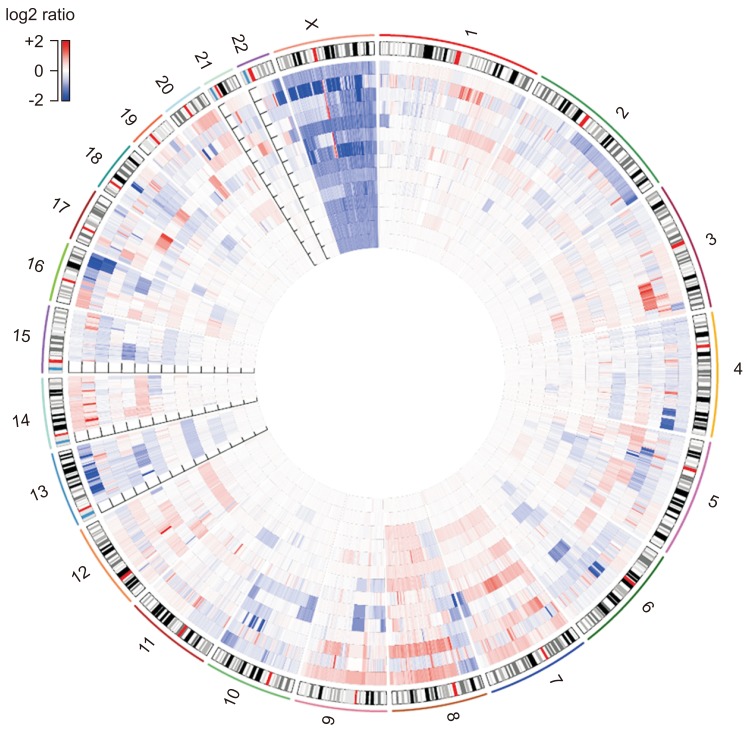
Genetic alterations were most frequently observed in the androgen receptor (AR) (42.9%, n=6/14), TP53 (14.3%, n=2/14), and PTEN (14.3%, n=2/14) genes in the Oncomine panel. AR amplification was the most common CNA (35.7%, n=5/14). As a result of LC-WGS, CNAs were confirmed in about 92.9% (n=13/14) of the samples in regions Xq12, 8q24.21, and 11q13.3 (gains) and in regions 6q16.1, 8p23.1, 10q25.1, 16q24.2, 18q12.3, Xq25, and Xq26.3 (losses). All CNAs identified in the Oncomine panel matched the results of LC-WGS. Additionally, LC-WGS of two plasma samples that matched tumor tissues revealed that CNA patterns of plasma samples (circulating tumor DNA) were very similar to those detected in tumor samples.
CONCLUSIONS
Our data showed that the characteristics of mutations and CNAs in Korean patients with advanced PCa were similar to those observed in previous studies.
MeSH Terms
Figure
Reference
-
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015; 65:87–108. PMID: 25651787.
Article2. Cooperberg MR, Chan JM. Epidemiology of prostate cancer. World J Urol. 2017; 35:849. PMID: 28509969.
Article3. Sartor O, de Bono JS. Metastatic prostate cancer. N Engl J Med. 2018; 378:1653–1654.
Article4. Ritch CR, Cookson MS. Advances in the management of castration resistant prostate cancer. BMJ. 2016; 355:i4405. PMID: 27754846.
Article5. Gandhi J, Afridi A, Vatsia S, Joshi G, Joshi G, Kaplan SA, et al. The molecular biology of prostate cancer: current understanding and clinical implications. Prostate Cancer Prostatic Dis. 2018; 21:22–36. PMID: 29282359.6. Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012; 487:239–243. PMID: 22722839.
Article7. Barbieri CE, Bangma CH, Bjartell A, Catto JW, Culig Z, Gronberg H, et al. The mutational landscape of prostate cancer. Eur Urol. 2013; 64:567–576. PMID: 23759327.
Article8. Cancer Genome Atlas Research N. The molecular taxonomy of primary prostate cancer. Cell. 2015; 163:1011–1025. PMID: 26544944.9. Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015; 161:1215–1228. PMID: 26000489.
Article10. Pritchard CC, Mateo J, Walsh MF, De Sarkar N, Abida W, Beltran H, et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med. 2016; 375:443–453. PMID: 27433846.
Article11. Mateo J, Carreira S, Sandhu S, Miranda S, Mossop H, Perez-Lopez R, et al. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med. 2015; 373:1697–1708. PMID: 26510020.
Article12. Koo KC, Lee KS, Chung BH. Urologic cancers in Korea. Jpn J Clin Oncol. 2015; 45:805–811. PMID: 26117494.
Article13. Hussain MH. The role of genomics in patients with advanced prostate cancer. Clin Adv Hematol Oncol. 2017; 15:770–772. PMID: 29040256.14. Quigley DA, Dang HX, Zhao SG, Lloyd P, Aggarwal R, Alumkal JJ, et al. Genomic hallmarks and structural variation in metastatic prostate cancer. Cell. 2018; 174:758–769.e9. PMID: 30033370.
Article15. Gao T, Mei Y, Sun H, Nie Z, Liu X, Wang S. The association of Phosphatase and tensin homolog (PTEN) deletion and prostate cancer risk: a meta-analysis. Biomed Pharmacother. 2016; 83:114–121. PMID: 27470558.
Article16. Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer [Erratum]. Cell. 2015; 162:454. PMID: 28843286.17. Wyatt AW, Annala M, Aggarwal R, Beja K, Feng F, Youngren J, et al. Concordance of circulating tumor DNA and matched metastatic tissue biopsy in prostate cancer. J Natl Cancer Inst. 2017; 109.
Article18. Annala M, Vandekerkhove G, Khalaf D, Taavitsainen S, Beja K, Warner EW, et al. Circulating tumor DNA genomics correlate with resistance to abiraterone and enzalutamide in prostate cancer. Cancer Discov. 2018; 8:444–457. PMID: 29367197.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Utility of Next-Generation Sequencing for Deciphering Intratumor Heterogeneity in Prostate Cancer
- Next-Generation Sequencing in Prostate Cancer
- Target-Enhanced Whole-Genome Sequencing Shows Clinical Validity Equivalent to Commercially Available Targeted Oncology Panel
- Trends in Next-Generation Sequencing and a New Era for Whole Genome Sequencing
- Validation of Customized Cancer Panel for Detecting Somatic Mutations and Copy Number Alterations