Lab Med Online.
2019 Apr;9(2):37-44. 10.3343/lmo.2019.9.2.37.
Essential Elements for Establishing Clinical Next-generation Sequencing Testing
- Affiliations
-
- 1Department of Laboratory Medicine and Biomedical Research Institute, Pusan National University Yangsan Hospital, Yangsan, Korea.
- 2Department of Laboratory Medicine, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea. wlee1@amc.seoul.kr
- KMID: 2441822
- DOI: http://doi.org/10.3343/lmo.2019.9.2.37
Abstract
- Over the past decade, next-generation sequencing (NGS) has evolved at an astonishing pace and has revolutionized clinical medicine as well as genomics research. The rapid advancements in NGS technologies have been accompanied by accumulating evidence of the analytical and clinical validity, and clinical utility of NGS. NGS is used worldwide. This review provides medical technicians and laboratory physicians with the essential elements for establishing clinical NGS testing. Here the authors briefly describe the advantages and drawbacks of currently available NGS platforms, potential sources of error in NGS workflow, and reference materials.
MeSH Terms
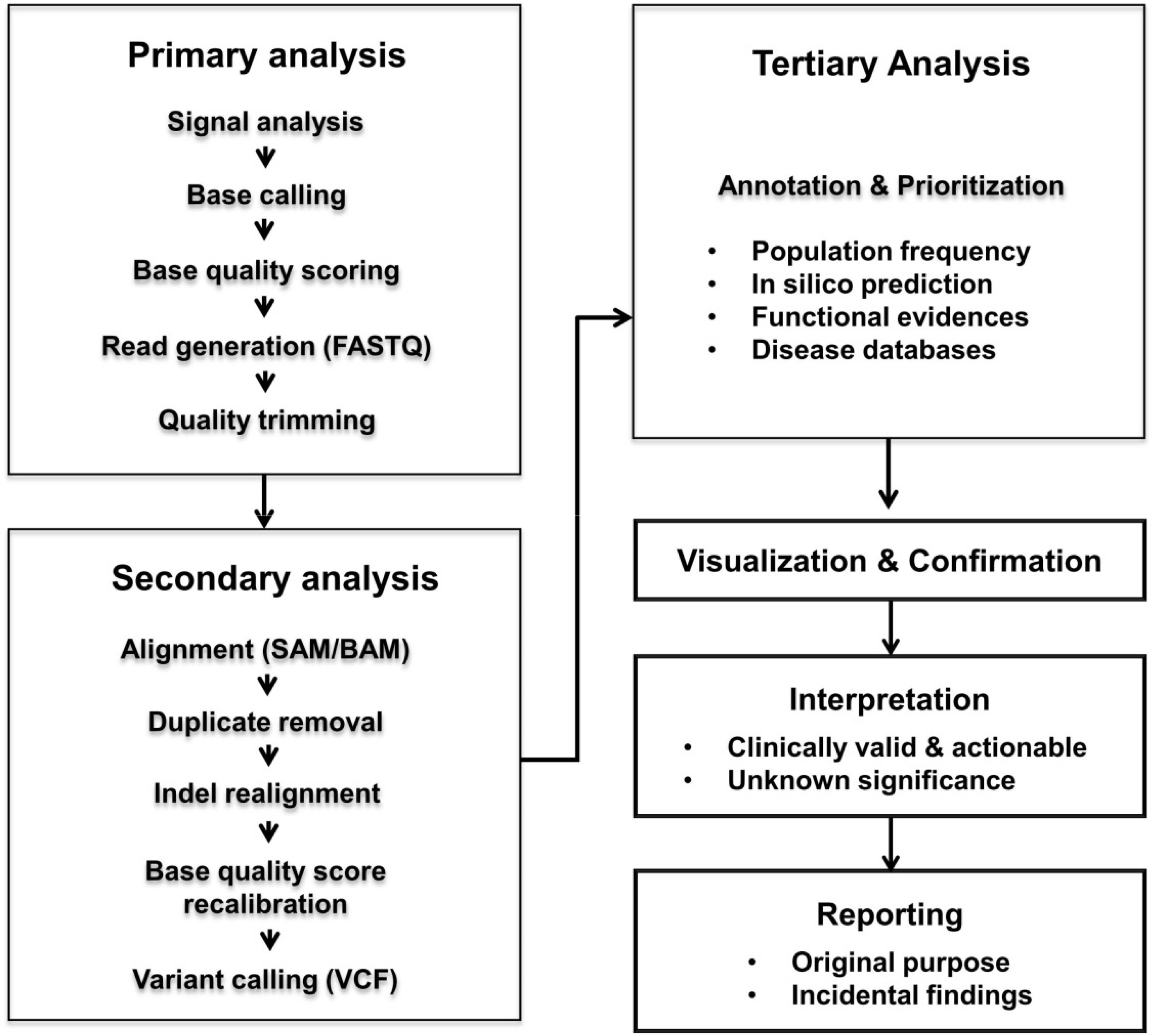
Figure
Reference
-
References
1. Levy SE and Myers RM. Advancements in next-generation sequencing. Annu Rev Genomics Hum Genet. 2016; 17:95–115.2. Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016; 17:333–51.
Article3. Minear MA, Alessi S, Allyse M, Michie M, Chandrasekharan S. Noninvasive prenatal genetic testing: current and emerging ethical, legal, and social issues. Annu Rev Genomics Hum Genet. 2015; 16:369–98.
Article4. Park KJ, Park S, Lee E, Park JH, Park JH, Park HD, et al. A population-based genomic study of inherited metabolic diseases detected through newborn screening. Ann Lab Med. 2016; 36:561–72.5. Bodian DL, Klein E, Iyer RK, Wong WS, Kothiyal P, Stauffer D, et al. Utility of whole-genome sequencing for detection of newborn screening disorders in a population cohort of 1,696 neonates. Genet Med. 2016; 18:221–30.
Article6. Pritchard CC, Salipante SJ, Koehler K, Smith C, Scroggins S, Wood B, et al. Validation and implementation of targeted capture and sequencing for the detection of actionable mutation, copy number variation, and gene rearrangement in clinical cancer specimens. J Mol Diagn. 2014; 16:56–67.
Article7. Lih CJ, Sims DJ, Harrington RD, Polley EC, Zhao Y, Mehaffey MG, et al. Analytical validation and application of a targeted next-generation sequencing mutation-detection assay for use in treatment assignment in the NCI-MPACT trial. J Mol Diagn. 2016; 18:51–67.
Article8. Saunders CJ, Miller NA, Soden SE, Dinwiddie DL, Noll A, Alnadi NA, et al. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci Transl Med. 2012; 4:154ra135.
Article9. Segal JP. Next-generation profciency testing. J Mol Diagn. 2016; 18:469–70.10. Aziz N, Zhao Q, Bry L, Driscoll DK, Funke B, Gibson JS, et al. College of American Pathologists'laboratory standards for next-generation sequencing clinical tests. Arch Pathol Lab Med. 2015; 139:481–93.11. Aird D, Ross MG, Chen WS, Danielsson M, Fennell T, Russ C, et al. Analyzing and minimizing PCR amplifcation bias in Illumina sequencing libraries. Genome Biol. 2011; 12:R18.12. Benjamini Y and Speed TP. Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res. 2012; 40:e72.13. Jennings LJ, Arcila ME, Corless C, Kamel-Reid S, Lubin IM, Pfeifer J, et al. Guidelines for validation of next-generation sequencing-based oncology panels: a joint consensus recommendation of the Association for Molecular Pathology and College of American Pathologists. J Mol Diagn. 2017; 19:341–65.14. Robasky K, Lewis NE, Church GM. The role of replicates for error mitigation in next-generation sequencing. Nat Rev Genet. 2014; 15:56–62.
Article15. Jones S, Anagnostou V, Lytle K, Parpart-Li S, Nesselbush M, Riley DR, et al. Personalized genomic analyses for cancer mutation discovery and interpretation. Sci Transl Med. 2015; 7:283ra53.
Article16. Kebschull JM and Zador AM. Sources of PCR-induced distortions in high-throughput sequencing data sets. Nucleic Acids Res. 2015; 43:e143.
Article17. Illumina. https://support.illumina.com/content/dam/illumina-marketing/documents/products/other/miseq-overclustering-primer-770-2014-038.pdf. (updated on August 2017).18. Pabinger S, Dander A, Fischer M, Snajder R, Sperk M, Efremova M, et al. A survey of tools for variant analysis of next-generation genome sequencing data. Brief Bioinform. 2014; 15:256–78.
Article19. Oliver GR, Hart SN, Klee EW. Bioinformatics for clinical next generation sequencing. Clin Chem. 2015; 61:124–35.
Article20. Chiara M and Pavesi G. Evaluation of quality assessment protocols for high throughput genome resequencing data. Front Genet. 2017; 8:94.
Article21. Gargis AS, Kalman L, Bick DP, da Silva C, Dimmock DP, Funke BH, et al. Good laboratory practice for clinical next-generation sequencing informatics pipelines. Nat Biotechnol. 2015; 33:689–93.
Article22. Santani A, Murrell J, Funke B, Yu Z, Hegde M, Mao R, et al. Development and validation of targeted next-generation sequencing panels for detection of germline variants in inherited diseases. Arch Pathol Lab Med. 2017; 141:787–97.
Article23. Hardwick SA, Deveson IW, Mercer TR. Reference standards for next-generation sequencing. Nat Rev Genet. 2017; 18:473–84.
Article24. Schmitt MW, Kennedy SR, Salk JJ, Fox EJ, Hiatt JB, Loeb LA. Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci U S A. 2012; 109:14508–13.
Article25. Sims DJ, Harrington RD, Polley EC, Forbes TD, Mehaffey MG, McGregor PM 3rd, et al. Plasmid-based materials as multiplex quality controls and calibrators for clinical next-generation sequencing assays. J Mol.26. Kudalkar EM, Almontashiri NA, Huang C, Anekella B, Bowser M, Hynes E, et al. Multiplexed reference materials as controls for diagnostic next-generation sequencing: a pilot investigating applications for hypertrophic cardiomyopathy. J Mol Diagn. 2016; 18:882–9.27. Illumina. https://www.illumina.com/content/dam/illumina-marketing/documents/products/technotes/hiseq-phix-control-v3-technical-note.pdf. (updated on August 2017).28. Duncavage EJ, Abel HJ, Merker JD, Bodner JB, Zhao Q, Voelkerding KV, et al. A model study of in silico profciency testing for clinical next-generation sequencing. Arch Pathol Lab Med. 2016; 140:1085–91.29. Davies KD, Farooqi MS, Gruidl M, Hill CE, Woolworth-Hirschhorn J, Jones H, et al. Multi-institutional FASTQ fle exchange as a means of profciency testing for next-generation sequencing bioinformatics and variant interpretation. J Mol Diagn. 2016; 18:572–9.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Genetic tests by next-generation sequencing in children with developmental delay and/or intellectual disability
- What’s new in molecular genetic pathology 2021: solid tumors and NGS panel selection
- Performance Evaluation of BRCA1/2 Genetic Test Using Next-Generation Sequencing Based on Target Capture Method
- Recent Advances in the Clinical Application of Next-Generation Sequencing
- Status of Next-Generation Sequencing-Based Genetic Diagnosis in Hematologic Malignancies in Korea (2017-2018)