Intest Res.
2018 Oct;16(4):529-536. 10.5217/ir.2018.00050.
Development and diversity of lactic acid producing bacteria and bifidobacteria in healthy full term Indian infants from Himachal Pradesh
- Affiliations
-
- 1Department of Biotechnology and Bioinformatics, Jaypee University of Information Technology, Waknaghat, India.
- KMID: 2434154
- DOI: http://doi.org/10.5217/ir.2018.00050
Abstract
- BACKGROUND/AIMS
The initial microbial colonization is a crucial step for the healthy development of an infant. Previous studies from India reported the dominance of target microbial species among Indian infants without any analysis on the diversity of target groups. This is the first study from India with an objective to investigate the establishment and diversity of lactic acid producing bacteria (LAB) and bifidobacteria in vaginally delivered, full term, breastfed infants for the first 4 months after birth.
METHODS
Present study used polymerase chain reaction-denaturing gradient gel electrophoresis (PCR-DGGE) based sequence analysis of LAB and bifidobacteria in healthy infants. The results were used to compare the development and early colonization by LAB and bifidobacteria using diversity indices during the initial months of development of gut microbiota in infants.
RESULTS
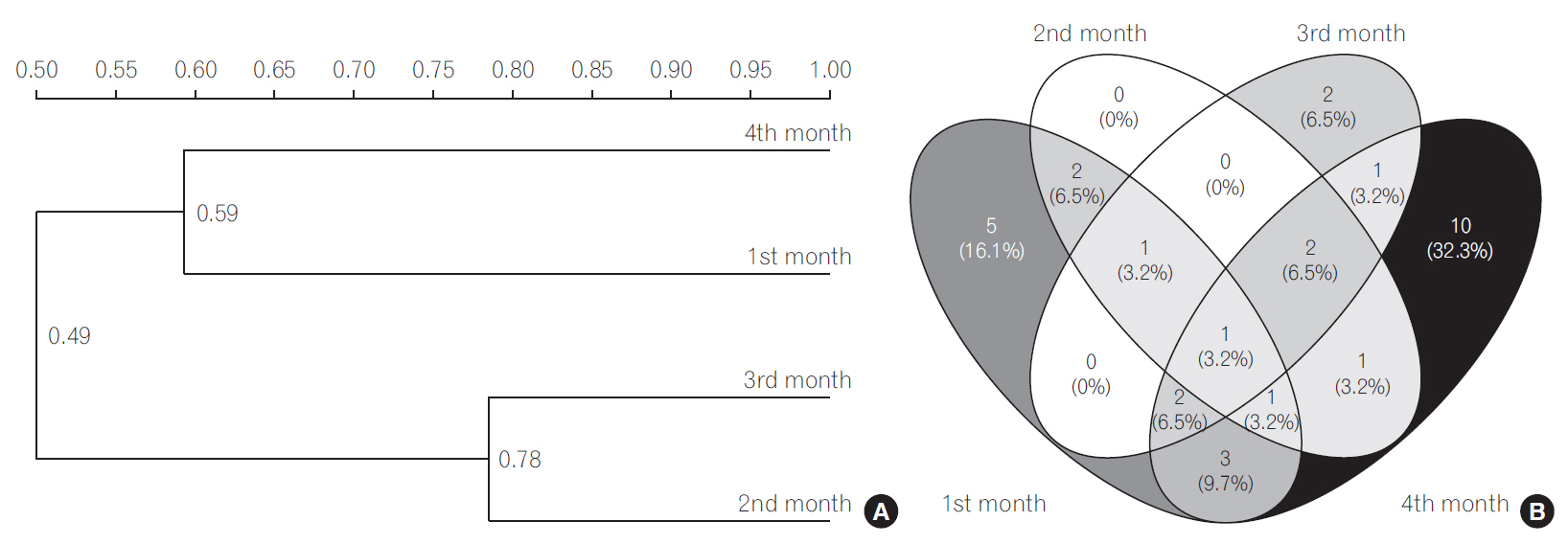
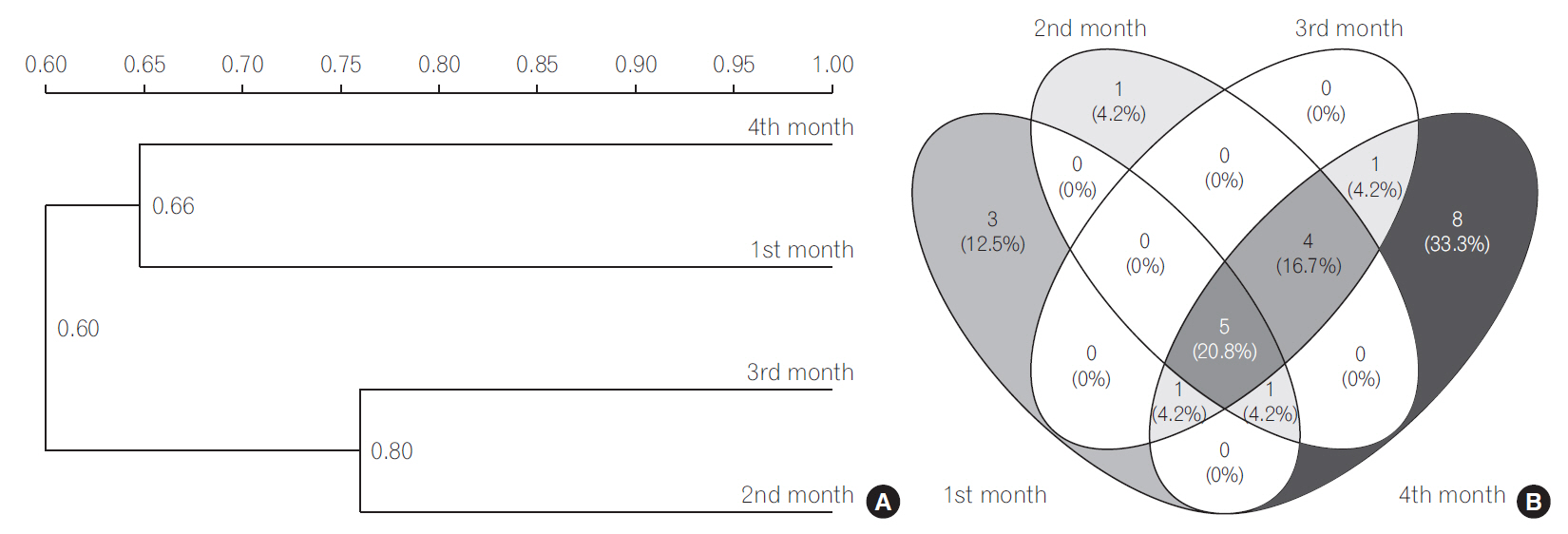
During the first 4 months, the Shannon diversity index (H) of LAB increased from 1.16 to 1.318 and for bifidobacteria the H increased from 0.975 to 1.293 (P < 0.05). Higher Sorenson's pair wise similarity coefficient was observed for LAB and bifidobacteria during 2nd and the 3rd month. The species of the genera Enterococcus, Streptococcus, and Lactobacillus were dominant among the LAB group whereas Bifidobacterium breve was dominant species among Bifidobacterium group.
CONCLUSIONS
Our results indicate that in breast fed infants, the microbial diversity of LAB and bifidobacteria increased during the period of study.
Keyword
MeSH Terms
Figure
Reference
-
1. Tan H, O’Toole PW. Impact of diet on the human intestinal microbiota. Curr Opin Food Sci. 2015; 2:71–77.
Article2. Jiménez E, Marín ML, Martín R, et al. Is meconium from healthy newborns actually sterile? Res Microbiol. 2008; 159:187–193.
Article3. Li M, Wang M, Donovan SM. Early development of the gut microbiome and immune-mediated childhood disorders. Semin Reprod Med. 2014; 32:74–86.
Article4. Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007; 449:811–818.
Article5. Adlerberth I, Wold AE. Establishment of the gut microbiota in Western infants. Acta Paediatr. 2009; 98:229–238.
Article6. Wang M, Monaco MH, Donovan SM. Impact of early gut microbiota on immune and metabolic development and function. Semin Fetal Neonatal Med. 2016; 21:380–387.
Article7. Patel K, Konduru K, Patra AK, Chandel DS, Panigrahi P. Trends and determinants of gastric bacterial colonization of preterm neonates in a NICU setting. PLoS One. 2015; 10:e0114664. doi: 10.1371/journal.pone.0114664.
Article8. Fei P, Li L, Cai X, et al. Differences in the biodiversity of the fecal microbiota of infants with rotaviral diarrhea and healthy infants. Jundishapur J Microbiol. 2016; 9:e32356. doi: 10.5812/jjm.32356.
Article9. The situation of children in India: a profile. United Nations Children’s Fund (UNICEF) Web site. https://www.unicef.org/sitan/files/SitAn_India_May_2011.pdf. Accessed Jun 12, 2018.10. Kabeerdoss J, Ferdous S, Balamurugan R, et al. Development of the gut microbiota in southern Indian infants from birth to 6 months: a molecular analysis. J Nutr Sci. 2013; 2:e18. doi: 10.1017/jns.2013.6.
Article11. Pandey PK, Siddharth J, Verma P, Bavdekar A, Patole MS, Shouche YS. Molecular typing of fecal eukaryotic microbiota of human infants and their respective mothers. J Biosci. 2012; 37:221–226.
Article12. Possemiers S, Verthé K, Uyttendaele S, Verstraete W. PCR-DGGE-based quantification of stability of the microbial community in a simulator of the human intestinal microbial ecosystem. FEMS Microbiol Ecol. 2004; 49:495–507.
Article13. Heilig HG, Zoetendal EG, Vaughan EE, Marteau P, Akkermans AD, de Vos WM. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl Environ Microbiol. 2002; 68:114–123.
Article14. Mangin I, Suau A, Magne F, et al. Characterization of human intestinal bifidobacteria using competitive PCR and PCR-TTGE. FEMS Microbiol Ecol. 2006; 55:28–37.
Article15. Mahajan R, Nikitina A, Nozhevnikova A, Goel G. Microbial diversity in an anaerobic digester with biogeographical proximity to geothermally active region. Environ Technol. 2016; 37:2694–2702.
Article16. Shannon CE, Weaver W. The mathematical theory of communication. Urbana: University Illinois Press;1963.17. Pielou EC. An introduction to mathematical ecology. New York: Wiley;1969.18. Schwiertz A, Gruhl B, Löbnitz M, Michel P, Radke M, Blaut M. Development of the intestinal bacterial composition in hospitalized preterm infants in comparison with breast-fed, full-term infants. Pediatr Res. 2003; 54:393–399.
Article19. Albesharat R, Ehrmann MA, Korakli M, Yazaji S, Vogel RF. Phenotypic and genotypic analyses of lactic acid bacteria in local fermented food, breast milk and faeces of mothers and their babies. Syst Appl Microbiol. 2011; 34:148–155.
Article20. Cong X, Xu W, Janton S, et al. Gut microbiome developmental patterns in early life of preterm infants: impacts of feeding and gender. PLoS One. 2016; 11:e0152751. doi: 10.1371/journal.pone.0152751.
Article21. von Wintersdorff CJ, Wolffs PF, Savelkoul PH, et al. The gut resistome is highly dynamic during the first months of life. Future Microbiol. 2016; 11:501–510.
Article22. Solís G, de Los Reyes-Gavilan CG, Fernández N, Margolles A, Gueimonde M. Establishment and development of lactic acid bacteria and bifidobacteria microbiota in breast-milk and the infant gut. Anaerobe. 2010; 16:307–310.
Article23. Castanys-Muñoz E, Martin MJ, Vazquez E. Building a beneficial microbiome from birth. Adv Nutr. 2016; 7:323–330.
Article24. Al-Kharousi ZS, Guizani N, Al-Sadi AM, Al-Bulushi IM, Shaharoona B. Hiding in fresh fruits and vegetables: opportunistic pathogens may cross geographical barriers. Int J Microbiol. 2016; 2016:4292417. doi: 10.1155/2016/4292417.
Article25. Jost T, Lacroix C, Braegger C, Chassard C. Assessment of bacterial diversity in breast milk using culture-dependent and culture-independent approaches. Br J Nutr. 2013; 110:1253–1262.
Article26. Songjinda P, Nakayama J, Kuroki Y, et al. Molecular monitoring of the developmental bacterial community in the gastrointestinal tract of Japanese infants. Biosci Biotechnol Biochem. 2005; 69:638–641.
Article27. Koort J, Coenye T, Vandamme P, Sukura A, Björkroth J. Enterococcus hermanniensis sp. nov., from modified-atmosphere-packaged broiler meat and canine tonsils. Int J Syst Evol Microbiol. 2004; 54(Pt 5):1823–1827.
Article28. Kubota H, Tsuji H, Matsuda K, Kurakawa T, Asahara T, Nomoto K. Detection of human intestinal catalase-negative, Gram-positive cocci by rRNA-targeted reverse transcription-PCR. Appl Environ Microbiol. 2010; 76:5440–5451.
Article29. Mahajan R, Nikitina A, Litti Y, Nozhevnikova A, Goel G. Autochthonous microbial community associated with pine needle forest litterfall influences its degradation under natural environmental conditions. Environ Monit Assess. 2016; 188:417. doi: 10.1007/s10661-016-5421-1.
Article30. Martín V, Maldonado-Barragán A, Moles L, et al. Sharing of bacterial strains between breast milk and infant feces. J Hum Lact. 2012; 28:36–44.
Article31. Mehanna NS, Tawfik NF, Salem MM, Effat BA, Gad El-Rab DA. Assessment of potential probiotic bacteria isolated from breast milk. Middle East J Sci Res. 2013; 14:354–360.32. Kharchenko NV, Cherdyntseva TA, Netrusov AI. New approaches for the isolation of bifidobacterial strains, their molecular characterization, and assessment of their probiotic potential. Microbiology. 2015; 84:419–424.
Article33. Vazquez-Gutierrez P, Lacroix C, Chassard C, Klumpp J, Jans C, Stevens MJ. Complete and assembled genome sequence of Bifidobacterium kashiwanohense PV20-2, isolated from the feces of an anemic kenyan infant. Genome Announc. 2015; 3:e01467–e01414. doi: 10.1128/genomeA.01467-14.
Article34. Delcenserie V, Loncaric D, Bonaparte C, et al. Bifidobacteria as indicators of faecal contamination along a sheep meat production chain. J Appl Microbiol. 2008; 104:276–284.
Article35. Yamamoto T, Zhou X, Williams CJ, Hochwalt A, Forney LJ. Bacterial populations in the vaginas of healthy adolescent women. J Pediatr Adolesc Gynecol. 2009; 22:11–18.
Article36. Gritz EC, Bhandari V. The human neonatal gut microbiome: a brief review. Front Pediatr. 2015; 3:17. doi: 10.3389/fped.2015.00017.
Article37. Urbaniak C, Angelini M, Gloor GB, Reid G. Human milk microbiota profiles in relation to birthing method, gestation and infant gender. Microbiome. 2016; 4:1. doi: 10.1186/s40168-015-0145-y.
Article38. Ruiz-Moyano S, Totten SM, Garrido DA, et al. Variation in consumption of human milk oligosaccharides by infant gut-associated strains of Bifidobacterium breve. Appl Environ Microbiol. 2013; 79:6040–6049.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Effects of Xylooligosaccharide Intake on Fecal Bifidobacteria,Lactic acid and Lipid Metabolism in Korean Young Women
- Hyperinflammatory syndrome in children during the coronavirus disease 2019 pandemic in sub-Himalayan region
- In vivo antitumor effects of lactic acid bacteria on sarcoma 180 and mouse lewis lung carcinoma
- Platelet Counts in Healthy Premature Infants
- A survey of research papers on the health benefits of kimchi and kimchi lactic acid bacteria