Ann Lab Med.
2019 May;39(3):327-329. 10.3343/alm.2019.39.3.327.
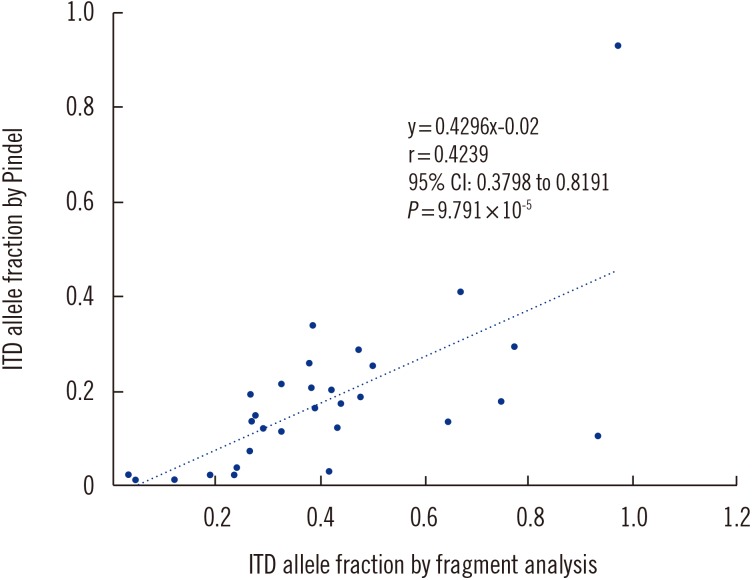
FLT3 Internal Tandem Duplication in Patients With Acute Myeloid Leukemia Is Readily Detectable in a Single Next-Generation Sequencing Assay Using the Pindel Algorithm
- Affiliations
-
- 1Department of Laboratory Medicine, Yonsei University College of Medicine, Seoul, Korea. cjr0606@yuhs.ac
- 2Department of Internal Medicine, Yonsei University College of Medicine, Seoul, Korea.
- KMID: 2431612
- DOI: http://doi.org/10.3343/alm.2019.39.3.327
Abstract
- No abstract available.
MeSH Terms
Figure
Cited by 1 articles
-
Phospholipase C Beta 2 Protein Overexpression Is a Favorable Prognostic Indicator in Newly Diagnosed Normal Karyotype Acute Myeloid Leukemia
Mi Suk Park, Young Eun Lee, Hye Ran Kim, Jong Hee Shin, Hyun Wook Cho, Jun Hyung Lee, Myung Geun Shin
Ann Lab Med. 2021;41(4):409-413. doi: 10.3343/alm.2021.41.4.409.
Reference
-
1. National Comprehensive Cancer Network. Acute Myeloid Leukemia (Version 2.2018). Updated on Aug 2018, registration required. https://www.nccn.org/professionals/physician_gls/pdf/aml.pdf.2. Chevallier P, Labopin M, Turlure P, Prebet T, Pigneux A, Hunault M, et al. A new Leukemia Prognostic Scoring System for refractory/relapsed adult acute myelogeneous leukaemia patients: a GOELAMS study. Leukemia. 2011; 25:939–944. PMID: 21331073.3. Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017; 129:424–447. PMID: 27895058.4. Degryse S, Cools J. JAK kinase inhibitors for the treatment of acute lymphoblastic leukemia. J Hematol Oncol. 2015; 8:91. PMID: 26208852.5. Wander SA, Levis MJ, Fathi AT. The evolving role of FLT3 inhibitors in acute myeloid leukemia: quizartinib and beyond. Ther Adv Hematol. 2014; 5:65–77. PMID: 24883179.6. Li H. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics. 2014; 30:2843–2851. PMID: 24974202.7. Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013; 31:213–219. PMID: 23396013.8. Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009; 25:2865–2871. PMID: 19561018.9. Zwaan CM, Meshinchi S, Radich JP, Veerman AJ, Huismans DR, Munske L, et al. FLT3 internal tandem duplication in 234 children with acute myeloid leukemia: prognostic significance and relation to cellular drug resistance. Blood. 2003; 102:2387–2394. PMID: 12816873.10. Spencer DH, Abel HJ, Lockwood CM, Payton JE, Szankasi P, Kelley TW, et al. Detection of FLT3 internal tandem duplication in targeted, short-read-length, next-generation sequencing data. J Mol Diagn. 2013; 15:81–93. PMID: 23159595.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Prognostic Significance of FLT3 Internal Tandem Duplication in Acute Myeloid Leukemia with Normal Karyotype
- FLT3 Internal Tandem Duplication in Acute Myeloid Leukemia with Normal Karyotype
- Prevalence of FLT3 Internal Tandem Duplication in Adult Acute Myelogenous Leukemia
- Molecular Risk Stratification using Next-generation Sequencing in Acute Myeloid Leukemia
- FLT3 mutations in acute myeloid leukemia: a review focusing on clinically applicable drugs