Ann Lab Med.
2017 Nov;37(6):475-483. 10.3343/alm.2017.37.6.475.
Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry in Clinical Microbiology: What Are the Current Issues?
- Affiliations
-
- 1Scientific Office, bioMérieux, La Balme Les Grottes, France. alex.vanbelkum@biomerieux.com
- 2Innovation Department, bioMérieux, La Balme Les Grottes, France.
- KMID: 2425987
- DOI: http://doi.org/10.3343/alm.2017.37.6.475
Abstract
- Matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) has revolutionized the identification of microbial species in clinical microbiology laboratories. MALDI-TOF-MS has swiftly become the new gold-standard method owing to its key advantages of simplicity and robustness. However, as with all new methods, adoption of the MALDI-TOF MS approach is still not widespread. Optimal sample preparation has not yet been achieved for several applications, and there are continuing discussions on the need for improved database quality and the inclusion of additional microbial species. New applications such as in the field of antimicrobial susceptibility testing have been proposed but not yet translated to the level of ease and reproducibility that one should expect in routine diagnostic systems. Finally, during routine identification testing, unexpected results are regularly obtained, and the best methods for transmitting these results into clinical care are still evolving. We here discuss the success of MALDI-TOF MS in clinical microbiology and highlight fields of application that are still amenable to improvement.
Keyword
MeSH Terms
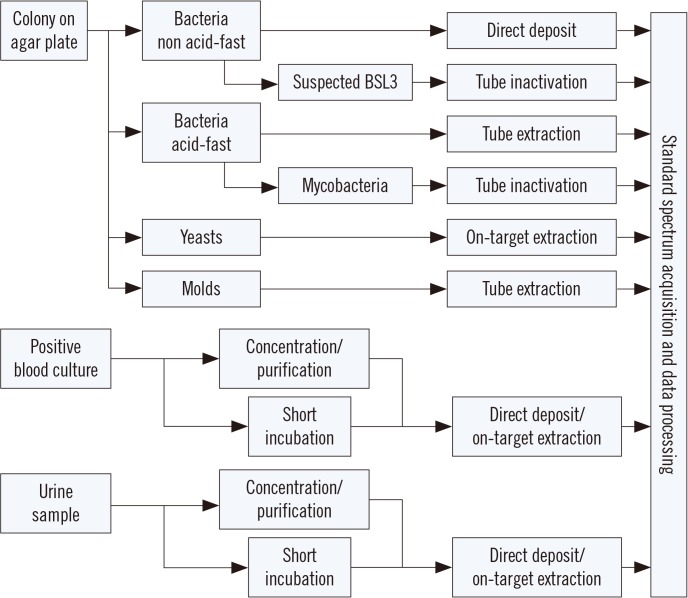
Figure
Cited by 1 articles
-
Performance Evaluation of VITEK MS for the Identification of a Wide Spectrum of Clinically Relevant Filamentous Fungi Using a Korean Collection
Ju Hyeon Shin, Soo Hyun Kim, Dain Lee, Seung Yeob Lee, Sejong Chun, Jun Hyung Lee, Eun Jeong Won, Hyun Jung Choi, Hyun Woo Choi, Seung Jung Kee, Myung Geun Shin, Jong Hee Shin
Ann Lab Med. 2021;41(2):214-220. doi: 10.3343/alm.2021.41.2.214.
Reference
-
1. van Belkum A, Welker M, Dunne WM Jr, Girard V. The infallible microbial identification test: does it exist? J Clin Microbiol. 2015; 53:1786. PMID: 25883325.2. Vandamme P, Peeters C. Time to revisit polyphasic taxonomy. Antonie Van Leeuwenhoek. 2014; 106:57–65. PMID: 24633913.3. Šedo O, Sedláček I, Zdráhal Z. Sample preparation methods for MALDI-MS profiling of bacteria. Mass Spectrom Rev. 2011; 30:417–434. PMID: 21500244.4. Lasch P, Jacob D, Grunow R, Schwecke T, Doellinger J. Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) for the identification of highly pathogenic bacteria. Trends Analyt Chem. 2016; 85:103–111.5. Dunne WM Jr, Doing K, Miller E, Miller E, Moreno E, Baghli M, et al. Rapid inactivation of Mycobacterium and Nocardia species before identification using matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2014; 52:3654–3659. PMID: 25078917.6. Mesureur J, Ranaldi S, Monnin V, Girard V, Arend S, Welker M, et al. A simple and safe protocol for preparing Brucella samples for matrix-assisted laser desorption/ionization-time of flight mass spectrometry analysis. J Clin Microbiol. 2016; 54:449–452. PMID: 26582837.7. Lasch P, Nattermann H, Erhard M, Stämmler M, Grunow R, Bannert N, et al. MALDI-TOF mass spectrometry compatible inactivation method for highly pathogenic microbial cells and spores. Anal Chem. 2008; 80:2026–2034. PMID: 18290666.8. Fothergill A, Kasinathan V, Hyman J, Walsh J, Drake T, Wang YF. Rapid identification of bacteria and yeasts from positive-blood-culture bottles by using a lysis-filtration method and matrix-assisted laser desorption ionization-time of flight mass spectrum analysis with the SARAMIS database. J Clin Microbiol. 2013; 51:805–809. PMID: 23254131.9. Nonnemann B, Tvede M, Bjarnsholt T. Identification of pathogenic microorganisms directly from positive blood vials by matrix-assisted laser desorption/ionization time of flight mass spectrometry. Apmis. 2013; 121:871–877. PMID: 23331371.10. Leli C, Cenci E, Cardaccia A, Moretti A, D'Alò F, Pagliochini R, et al. Rapid identification of bacterial and fungal pathogens from positive blood cultures by MALDI-TOF MS. Int J Med Microbiol. 2013; 303:205–209. PMID: 23602511.11. Köck R, Wüllenweber J, Horn D, Lanckohr C, Becker K, Idelevich EA. Implementation of short incubation MALDI-TOF MS identification from positive blood cultures in routine diagnostics and effects on empiric antimicrobial therapy. Antimicrob Resist Infect Control. 2017; 6:12. PMID: 28101334.12. Altun O, Botero-Kleiven S, Carlsson S, Ullberg M, Özenci V. Rapid identification of bacteria from positive blood culture bottles by MALDI-TOF MS following short-term incubation on solid media. J Med Microbiol. 2015; 64:1346–1352. PMID: 26361761.13. Curtoni A, Cipriani R, Marra ES, Barbui AM, Cavallo R, Costa C. Rapid identification of microorganisms from positive blood culture by MALDI-TOF MS after short-term incubation on solid medium. Curr Microbiol. 2017; 74:97–102. PMID: 27858149.14. Angeletti S, Dicuonzo G, D'Agostino A, Avola A, Crea F, Palazzo C, et al. Turnaround time of positive blood cultures after the introduction of matrix-assisted laser desorption-ionization time-of-flight mass spectrometry. New Microbiol. 2015; 38:379–386. PMID: 26147149.15. Schneiderhan W, Grundt A, Wörner S, Findeisen P, Neumaier M. Work flow analysis of around-the-clock processing of blood culture samples and integrated MALDI-TOF mass spectrometry analysis for the diagnosis of bloodstream infections. Clin Chem. 2013; 59:1649–1656. PMID: 23881934.16. Hsieh SY, Tseng CL, Lee YS, Kuo AJ, Sun CF, Lin YH, et al. Highly efficient classification and identification of human pathogenic bacteria by MALDI-TOF MS. Mol Cell Proteomics. 2008; 7:448–456. PMID: 18045801.17. Köhling HL, Bittner A, Müller KD, Buer J, Becker M, Rübben H, et al. Direct identification of bacteria in urine samples by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and relevance of defensins as interfering factors. J Med Microbiol. 2012; 61:339–344. PMID: 22275503.18. Seng P, Abat C, Rolain JM, Colson P, Lagier JC, Gouriet F, et al. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2013; 51:2182–2194. PMID: 23637301.19. Szabados F, Anders A, Kaase M, Marlinghaus L, Gatermann SG, Teske W, et al. Late periprosthetic joint infection due to Staphylococcus lugdunensis identified by Matrix-Assisted Laser Desorption/Ionisation Time of Flight Mass Spectrometry: a case report and review of the literature. Case Rep Med. 2011; 2011:608919. PMID: 21776276.20. Rodríguez-Sánchez B, Marín M, Sánchez-Carrillo C, Cercenado E, Ruiz A, Rodríguez-Créixems M, et al. Improvement of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry identification of difficult-to-identify bacteria and its impact in the workflow of a clinical microbiology laboratory. Diagn Microbiol Infect Dis. 2014; 79:1–6. PMID: 24602850.21. Welker M, Moore ER. Applications of whole-cell matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry in systematic microbiology. Syst Appl Microbiol. 2011; 34:2–11. PMID: 21288677.22. Lagier JC, Khelaifia S, Alou MT, Ndongo S, Dione N, Hugon P, et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016; 1:16203. PMID: 27819657.23. Rajilić-Stojanović M, de Vos WM. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev. 2014; 38:996–1047. PMID: 24861948.24. Grenfell RC, da Silva Junior AR, Del Negro GM, Munhoz RB, Gimenes VM, Assis DM, et al. Identification of Candida haemulonii complex species: Use of ClinProTools(TM) to overcome limitations of the Bruker Biotyper(TM), VITEK MS(TM) IVD, and VITEK MS(TM) RUO Databases. Front Microbiol. 2016; 7:940. PMID: 27379069.25. Lamoth F. Aspergillus fumigatus-related species in clinical practice. Front Microbiol. 2016; 7:683. PMID: 27242710.26. Lambiase A, Del Pezzo M, Cerbone D, Raia V, Rossano F, Catania MR. Rapid identification of Burkholderia cepacia complex species recovered from cystic fibrosis patients using matrix-assisted laser desorption ionization time-of-flight mass spectrometry. J Microbiol Methods. 2013; 92:145–149. PMID: 23201483.27. Han MS, Kim H, Lee Y, Kim M, Ku NS, Choi JY, et al. Relative prevalence and antimicrobial susceptibility of clinical isolates of Elizabethkingia species based on 16S rRNA gene sequencing. J Clin Microbiol. 2016; 55:274–280. PMID: 27847376.28. Fonseca EL, Ramos ND, Andrade BG, Morais LL, Marin MF, Vicente AC. A one-step multiplex PCR to identify Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae in the clinical routine. Diagn Microbiol Infect Dis. 2017; 87:315–317. PMID: 28139276.29. Teixeira AB, Barin J, Hermes DM, Barth AL, Martins AF. PCR assay based on the gyrB gene for rapid identification of Acinetobacter baumannii-calcoaceticus complex at species level. J Clin Lab Anal. 2017; 31:e22046.30. Marí-Almirall M, Cosgaya C, Higgins PG, Van Assche A, Telli M, Huys G, et al. MALDI-TOF/MS identification of species from the Acinetobacter baumannii (Ab) group revisited: inclusion of the novel A. seifertii and A. dijkshoorniae species. Clin Microbiol Infect. 2017; 23:210. PMID: 27919649.31. Rosselló-Móra R, Amann R. Past and future species definitions for Bacteria and Archaea. Syst Appl Microbiol. 2015; 38:209–216. PMID: 25747618.32. Whitman WB. Genome sequences as the type material for taxonomic descriptions of prokaryotes. Syst Appl Microbiol. 2015; 38:217–222. PMID: 25769508.33. Tortoli E, Richter E, Borroni E, Cabibbe AM, Capitolo E, Cittaro D, et al. Mycobacterium alsense sp. nov., a scotochromogenic slow grower isolated from clinical respiratory specimens. Int J Syst Evol Microbiol. 2016; 66:450–456. PMID: 26545358.34. Shahraki AH, Çavusoğlu C, Borroni E, Heidarieh P, Koksalan OK, Cabibbe AM, et al. Mycobacterium celeriflavum sp. nov., a rapidly growing scotochromogenic bacterium isolated from clinical specimens. Int J Syst Evol Microbiol. 2015; 65:510–515. PMID: 25389151.35. Nogueira CL, Whipps CM, Matsumoto CK, Chimara E, Droz S, Tortoli E, et al. Mycobacterium saopaulense sp. nov., a rapidly growing mycobacterium closely related to members of the Mycobacterium chelonae--Mycobacterium abscessus group. Int J Syst Evol Microbiol. 2015; 65:4403–4409. PMID: 26358475.36. Josten M, Dischinger J, Szekat C, Reif M, Al-Sabti N, Sahl HG, et al. Identification of agr-positive methicillin-resistant Staphylococcus aureus harbouring the class A mec complex by MALDI-TOF mass spectrometry. Int J Med Microbiol. 2014; 304:1018–1023. PMID: 25116838.37. Rhoads DD, Wang H, Karichu J, Richter SS. The presence of a single MALDI-TOF mass spectral peak predicts methicillin resistance in staphylococci. Diagn Microbiol Infect Dis. 2016; 86:257–261. PMID: 27568365.38. Fenyvesi VS, Urbán E, Bartha N, Abrók M, Kostrzewa M, Nagy E, et al. Use of MALDI-TOF/MS for routine detection of cfiA gene-positive Bacteroides fragilis strains. Int J Antimicrob Agents. 2014; 44:474–475. PMID: 25216544.39. Nagy E, Becker S, Sóki J, Urbán E, Kostrzewa M. Differentiation of division I (cfiA-negative) and division II (cfiA-positive) Bacteroides fragilis strains by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J Med Microbiol. 2011; 60:1584–1590. PMID: 21680764.40. Sparbier K, Schubert S, Kostrzewa M. MBT-ASTRA: A suitable tool for fast antibiotic susceptibility testing? Methods. 2016; 104:48–54. PMID: 26804565.41. Jung JS, Hamacher C, Gross B, Sparbier K, Lange C, Kostrzewa M, et al. Evaluation of a semiquantitative matrix-assisted laser desorption ionization-time of flight mass spectrometry method for rapid antimicrobial susceptibility testing of positive blood cultures. J Clin Microbiol. 2016; 54:2820–2824. PMID: 27629893.42. Sparbier K, Lange C, Jung J, Wieser A, Schubert S, Kostrzewa M. MALDI biotyper-based rapid resistance detection by stable-isotope labeling. J Clin Microbiol. 2013; 51:3741–3748. PMID: 24006001.43. Wieser A, Schubert S. MALDI-TOF MS entering the microbiological diagnostic laboratory - from fast identification to resistance testing. TrAC Trends Anal Chem. 2016; 84:80–87.44. Hrabák J, Walková R, Studentová V, Chudácková E, Bergerová T. Carbapenemase activity detection by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2011; 49:3222–3227. PMID: 21775535.45. Hooff GP, van Kampen JJ, Meesters RJ, van Belkum A, Goessens WH, Luider TM. Characterization of β-lactamase enzyme activity in bacterial lysates using MALDI-mass spectrometry. J Proteome Res. 2012; 11:79–84. PMID: 22013912.46. Sparbier K, Schubert S, Weller U, Boogen C, Kostrzewa M. Matrix-assisted laser desorption ionization-time of flight mass spectrometry-based functional assay for rapid detection of resistance against β-lactam antibiotics. J Clin Microbiol. 2012; 50:927–937. PMID: 22205812.47. Hrabák J, Studentová V, Walková R, Zemlicková H, Jakubu V, Chudácková E, et al. Detection of NDM-1, VIM-1, KPC, OXA-48, and OXA-162 carbapenemases by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2012; 50:2441–2443. PMID: 22553235.48. Knox J, Jadhav S, Sevior D, Agyekum A, Whipp M, Waring L, et al. Phenotypic detection of carbapenemase-producing Enterobacteriaceae by use of matrix-assisted laser desorption ionization-time of flight mass spectrometry and the Carba NP test. J Clin Microbiol. 2014; 52:4075–4077. PMID: 25187633.49. Chong PM, McCorrister SJ, Unger MS, Boyd DA, Mulvey MR, Westmacott GR. MALDI-TOF MS detection of carbapenemase activity in clinical isolates of Enterobacteriaceae spp., Pseudomonas aeruginosa, and Acinetobacter baumannii compared against the Carba-NP assay. J Microbiol Methods. 2015; 111:21–23. PMID: 25644285.50. Carvalhaes CG, da Silva AC, Streling AP, Cayô R, Rockstroh AC, Machado AM, et al. Detection of carbapenemase activity using VITEK MS: interplay of carbapenemase type and period of incubation. J Med Microbiol. 2015; 64:946–947. PMID: 26293667.51. Carvalhaes CG, Cayô R, Visconde MF, Barone T, Frigatto EA, Okamoto D, et al. Detection of carbapenemase activity directly from blood culture vials using MALDI-TOF MS: a quick answer for the right decision. J Antimicrob Chemother. 2014; 69:2132–2136. PMID: 24722840.52. Johansson Å, Nagy E, Sóki J. Instant screening and verification of carbapenemase activity in Bacteroides fragilis in positive blood culture, using matrix-assisted laser desorption ionization--time of flight mass spectrometry. J Med Microbiol. 2014; 63:1105–1110. PMID: 24850880.53. Kulkarni MV, Zurita AN, Pyka JS, Murray TS, Hodsdon ME, Peaper DR. Use of imipenem to detect KPC, NDM, OXA, IMP, and VIM carbapenemase activity from gram-negative rods in 75 minutes using liquid chromatography-tandem mass spectrometry. J Clin Microbiol. 2014; 52:2500–2505. PMID: 24789180.54. Peaper DR, Kulkarni MV, Tichy AN, Jarvis M, Murray TS, Hodsdon ME. Rapid detesction of carbapenemase activity through monitoring ertapenem hydrolysis in Enterobacteriaceae with LC-MS/MS. Bioanalysis. 2013; 5:147–157. PMID: 23330558.55. Fleurbaaij F, Heemskerk AA, Russcher A, Klychnikov OI, Deelder AM, Mayboroda OA, et al. Capillary-electrophoresis mass spectrometry for the detection of carbapenemases in (multi-)drug-resistant Gram-negative bacteria. Anal Chem. 2014; 86:9154–9161. PMID: 25155175.56. Hart PJ, Wey E, McHugh TD, Balakrishnan I, Belgacem O. A method for the detection of antibiotic resistance markers in clinical strains of Escherichia coli using MALDI mass spectrometry. J Microbiol Methods. 2015; 111:1–8. PMID: 25633625.57. Charretier Y, Dauwalder O, Franceschi C, Degout-Charmette E, Zambardi G, Cecchini T, et al. Rapid bacterial identification, resistance, virulence and type profiling using selected reaction monitoring mass spectrometry. Sci Rep. 2015; 5:13944. PMID: 26350205.58. Karlsson R, Gonzales-Siles L, Boulund F, Svensson-Stadler L, Skovbjerg S, Karlsson A, et al. Proteotyping: Proteomic characterization, classification and identification of microorganisms - A prospectus. Syst Appl Microbiol. 2015; 38:246–257. PMID: 25933927.59. Aguirre-Quiñonero A, Martínez-Martínez L. Non-molecular detection of carbapenemases in Enterobacteriaceae clinical isolates. J Infect Chemother. 2017; 23:1–11. PMID: 27769646.60. Monteferrante CG, Sultan S, Ten Kate MT, Dekker LJ, Sparbier K, Peer M, et al. Evaluation of different pretreatment protocols to detect accurately clinical carbapenemase-producing Enterobacteriaceae by MALDI-TOF. J Antimicrob Chemother. 2016; 71:2856–2867. PMID: 27287232.61. Mirande C, Canard I, Buffet Croix, Charrier JP, van Belkum A, Welker M, et al. Rapid detection of carbapenemase activity: benefits and weaknesses of MALDI-TOF MS. Eur J Clin Microbiol Infect Dis. 2015; 34:2225–2234. PMID: 26337432.62. van Wuijckhuijse AL, Stowers MA, Kleefsman WA, van Baar BLM, Kientz CE, Marijnissen JCM. Matrix-assisted laser desorption/ionisation aerosol time-of-flight mass spectrometry for the analysis of bioaerosols: development of a fast detector for airborne biological pathogens. J Aerosol Sci. 2005; 36:677–687.63. Madonna AJ, Basile F, Furlong E, Voorhees KJ. Detection of bacteria from biological mixtures using immunomagnetic separation combined with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2001; 15:1068–1074. PMID: 11404843.64. Ochoa ML, Harrington PB. Immunomagnetic isolation of enterohemorrhagic Escherichia coli O157:H7 from ground beef and identification by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and database searches. Anal Chem. 2005; 77:5258–5267. PMID: 16097767.65. Khot PD, Fisher MA. Novel approach for differentiating Shigella species and Escherichia coli by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2013; 51:3711–3716. PMID: 23985919.66. Paauw A, Jonker D, Roeselers G, Heng JM, Mars-Groenendijk RH, Trip H, et al. Rapid and reliable discrimination between Shigella species and Escherichia coli using MALDI-TOF mass spectrometry. Int J Med Microbiol. 2015; 305:446–452. PMID: 25912807.67. Pupo GM, Lan R, Reeves PR. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc Natl Acad Sci U S A. 2000; 97:10567–10572. PMID: 10954745.68. Buckwalter SP, Olson SL, Connelly BJ, Lucas BC, Rodning AA, Walchak RC, et al. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of Mycobacterium species, Nocardia species, and other aerobic Actinomycetes. J Clin Microbiol. 2016; 54:376–384. PMID: 26637381.69. Girard V, Mailler S, Welker M, Arsac M, Cellière B, Cotte-Pattat PJ, et al. Identification of mycobacterium spp. and nocardia spp. from solid and liquid cultures by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS). Diagn Microbiol Infect Dis. 2016; 86:277–283. PMID: 27567285.70. Patel R. Matrix-assisted laser desorption ionization-time of flight mass spectrometry in clinical microbiology. Clin Infect Dis. 2013; 57:564–572. PMID: 23595835.71. Tran A, Alby K, Kerr A, Jones M, Gilligan PH. Cost savings realized by implementation of routine microbiological identification by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2015; 53:2473–2479. PMID: 25994167.72. Tan KE, Ellis BC, Lee R, Stamper PD, Zhang SX, Carroll KC. Prospective evaluation of a matrix-assisted laser desorption ionization-time of flight mass spectrometry system in a hospital clinical microbiology laboratory for identification of bacteria and yeasts: a bench-by-bench study for assessing the impact on time to identification and cost-effectiveness. J Clin Microbiol. 2012; 50:3301–3308. PMID: 22855510.73. Neville SA, Lecordier A, Ziochos H, Chater MJ, Gosbell IB, Maley MW, et al. Utility of matrix-assisted laser desorption ionization-time of flight mass spectrometry following introduction for routine laboratory bacterial identification. J Clin Microbiol. 2011; 49:2980–2984. PMID: 21632894.74. Becker K, Heilmann C, Peters G. Coagulase-negative staphylococci. Clin Microbiol Rev. 2014; 27:870–926. PMID: 25278577.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- MALDI-TOF-MS Fingerprinting Provides Evidence of Urosepsis caused by Aerococcus urinae
- Application of Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Imaging Mass Spectrometry (MALDI-TOF IMS) for Premalignant Gastrointestinal Lesions
- Risk of inaccurate species identification by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and of false carbapenem resistance by automated susceptibility analysis of Enterobacter spp.
- Two Cases of Campylobacter jejuni Bacteremia from Patients with Diarrhea
- Reliability of Acinetobacter baumannii Identification with Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry