Microbiome in the Gut-Skin Axis in Atopic Dermatitis
- Affiliations
-
- 1Department of Pediatrics, Childhood Asthma Atopy Center, Environmental Health Center, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea. sjhong@amc.seoul.kr
- 2Department of Pediatrics, Chonnam National University Hospital, Chonnam National University Medical School, Gwangju, Korea.
- 3Asan Institute for Life Science, University of Ulsan College of Medicine, Seoul, Korea.
- KMID: 2421669
- DOI: http://doi.org/10.4168/aair.2018.10.4.354
Abstract
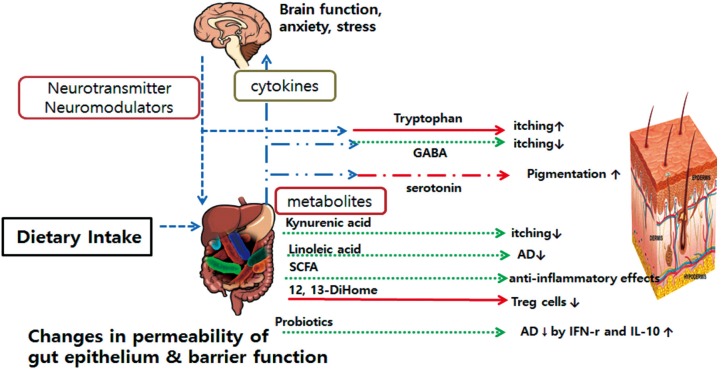
- The microbiome is vital for immune system development and homeostasis. Changes in microbial composition and function, termed dysbiosis, in the skin and the gut have recently been linked to alterations in immune responses and to the development of skin diseases, such as atopic dermatitis (AD). In this review, we summarize the recent findings on the gut and skin microbiome, highlighting the roles of major commensals in modulating skin and systemic immunity in AD. Although our understanding of the gut-skin axis is only beginning, emerging evidence indicates that the gut and skin microbiome could be manipulated to treat AD.
Keyword
MeSH Terms
Figure
Cited by 3 articles
-
Effects of
Lactobacillus pentosus in Children with Allergen-Sensitized Atopic Dermatitis
So Hyun Ahn, Wonsuck Yoon, So Young Lee, Hee Soon Shin, Mi Young Lim, Young-Do Nam, Young Yoo
J Korean Med Sci. 2020;35(18):e128. doi: 10.3346/jkms.2020.35.e128.Antibiotics-Induced Dysbiosis of Intestinal Microbiota Aggravates Atopic Dermatitis in Mice by Altered Short-Chain Fatty Acids
Ha-Jung Kim, Seung-Hwa Lee, Soo-Jong Hong
Allergy Asthma Immunol Res. 2020;12(1):137-148. doi: 10.4168/aair.2020.12.1.137.Imbalance of Gut Streptococcus, Clostridium, and Akkermansia Determines the Natural Course of Atopic Dermatitis in Infant
Yoon Mee Park, So-Yeon Lee, Mi-Jin Kang, Bong-Soo Kim, Min-Jung Lee, Sung Su Jung, Ji Sun Yoon, Hyun-Ju Cho, Eun Lee, Song-I Yang, Ju-Hee Seo, Hyo-Bin Kim, Dong In Suh, Youn Ho Shin, Kyung Won Kim, Kangmo Ahn, Soo-Jong Hong
Allergy Asthma Immunol Res. 2020;12(2):322-337. doi: 10.4168/aair.2020.12.2.322.
Reference
-
1. Dharmage SC, Lowe AJ, Matheson MC, Burgess JA, Allen KJ, Abramson MJ. Atopic dermatitis and the atopic march revisited. Allergy. 2014; 69:17–27. PMID: 24117677.
Article2. Flohr C, Mann J. New insights into the epidemiology of childhood atopic dermatitis. Allergy. 2014; 69:3–16. PMID: 24417229.
Article3. Lee SY, Yu J, Ahn KM, Kim KW, Shin YH, Lee KS, et al. Additive effect between IL-13 polymorphism and cesarean section delivery/prenatal antibiotics use on atopic dermatitis: a birth cohort study (COCOA). PLoS One. 2014; 9:e96603. PMID: 24848505.
Article4. Lee JY, Seo JH, Kwon JW, Yu J, Kim BJ, Lee SY, et al. Exposure to gene-environment interactions before 1 year of age may favor the development of atopic dermatitis. Int Arch Allergy Immunol. 2012; 157:363–371. PMID: 22123373.
Article5. Kim BJ, Lee SY, Kim HB, Lee E, Hong SJ. Environmental changes, microbiota, and allergic diseases. Allergy Asthma Immunol Res. 2014; 6:389–400. PMID: 25228995.
Article6. Al-Asmakh M, Zadjali F. Use of germ-free animal models in microbiota-related research. J Microbiol Biotechnol. 2015; 25:1583–1588. PMID: 26032361.
Article7. Lebeer S, Vanderleyden J, De Keersmaecker SC. Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat Rev Microbiol. 2010; 8:171–184. PMID: 20157338.
Article8. Ueno N, Fujiya M, Segawa S, Nata T, Moriichi K, Tanabe H, et al. Heat-killed body of lactobacillus brevis SBC8803 ameliorates intestinal injury in a murine model of colitis by enhancing the intestinal barrier function. Inflamm Bowel Dis. 2011; 17:2235–2250. PMID: 21987297.
Article9. Peng Y, Li A, Yu L, Qin G. The role of probiotics in prevention and treatment for patients with allergic rhinitis: a systematic review. Am J Rhinol Allergy. 2015; 29:292–298. PMID: 26163249.
Article10. Kim SO, Ah YM, Yu YM, Choi KH, Shin WG, Lee JY. Effects of probiotics for the treatment of atopic dermatitis: a meta-analysis of randomized controlled trials. Ann Allergy Asthma Immunol. 2014; 113:217–226. PMID: 24954372.
Article11. Rather IA, Bajpai VK, Kumar S, Lim J, Paek WK, Park YH. Probiotics and atopic dermatitis: an overview. Front Microbiol. 2016; 7:507. PMID: 27148196.
Article12. Levkovich T, Poutahidis T, Smillie C, Varian BJ, Ibrahim YM, Lakritz JR, et al. Probiotic bacteria induce a ‘glow of health’. PLoS One. 2013; 8:e53867. PMID: 23342023.
Article13. Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012; 336:489–493. PMID: 22442383.
Article14. Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013; 504:451–455. PMID: 24226773.
Article15. Zeng MY, Inohara N, Nuñez G. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 2017; 10:18–26. PMID: 27554295.
Article16. Gensollen T, Blumberg RS. Correlation between early-life regulation of the immune system by microbiota and allergy development. J Allergy Clin Immunol. 2017; 139:1084–1091. PMID: 28390575.
Article17. Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, Jenmalm MC. Low diversity of the gut microbiota in infants with atopic eczema. J Allergy Clin Immunol. 2012; 129:434–440. 440.e1–440.e2. PMID: 22153774.
Article18. Penders J, Stobberingh EE, Thijs C, Adams H, Vink C, van Ree R, et al. Molecular fingerprinting of the intestinal microbiota of infants in whom atopic eczema was or was not developing. Clin Exp Allergy. 2006; 36:1602–1608. PMID: 17177684.
Article19. Lee E, Lee SY, Kang MJ, Kim K, Won S, Kim BJ, et al. Clostridia in the gut and onset of atopic dermatitis via eosinophilic inflammation. Ann Allergy Asthma Immunol. 2016; 117:91–92.e1. PMID: 27179583.
Article20. Kirjavainen PV, Arvola T, Salminen SJ, Isolauri E. Aberrant composition of gut microbiota of allergic infants: a target of bifidobacterial therapy at weaning? Gut. 2002; 51:51–55. PMID: 12077091.
Article21. Nylund L, Nermes M, Isolauri E, Salminen S, de Vos WM, Satokari R. Severity of atopic disease inversely correlates with intestinal microbiota diversity and butyrate-producing bacteria. Allergy. 2015; 70:241–244. PMID: 25413686.
Article22. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013; 341:569–573. PMID: 23828891.23. Song H, Yoo Y, Hwang J, Na YC, Kim HS. Faecalibacterium prausnitzii subspecies-level dysbiosis in the human gut microbiome underlying atopic dermatitis. J Allergy Clin Immunol. 2016; 137:852–860. PMID: 26431583.24. Lee MJ, Kang MJ, Lee SY, Lee E, Kim K, Won S, et al. Perturbations of the gut microbiome genes in infants with atopic dermatitis according to feeding type. J Allergy Clin Immunol. 2018; 141:1310–1319. PMID: 29339259.25. Chang YS, Trivedi MK, Jha A, Lin YF, Dimaano L, García-Romero MT. Synbiotics for prevention and treatment of atopic dermatitis: a meta-analysis of randomized clinical trials. JAMA Pediatr. 2016; 170:236–242. PMID: 26810481.26. Huang YJ, Marsland BJ, Bunyavanich S, O'Mahony L, Leung DY, Muraro A, et al. The microbiome in allergic disease: current understanding and future opportunities-2017 PRACTALL document of the American Academy of Allergy, Asthma & Immunology and the European Academy of Allergy and Clinical Immunology. J Allergy Clin Immunol. 2017; 139:1099–1110. PMID: 28257972.27. Smits HH, Engering A, van der Kleij D, de Jong EC, Schipper K, van Capel TM, et al. Selective probiotic bacteria induce IL-10-producing regulatory T cells in vitro by modulating dendritic cell function through dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin. J Allergy Clin Immunol. 2005; 115:1260–1267. PMID: 15940144.28. Nowrouzian FL, Lina G, Hodille E, Lindberg E, Hesselmar B, Saalman R, et al. Superantigens and adhesins of infant gut commensal Staphylococcus aureus strains and association with subsequent development of atopic eczema. Br J Dermatol. 2017; 176:439–445. PMID: 27761891.29. Gueniche A, Philippe D, Bastien P, Reuteler G, Blum S, Castiel-Higounenc I, et al. Randomised double-blind placebo-controlled study of the effect of Lactobacillus paracasei NCC 2461 on skin reactivity. Benef Microbes. 2014; 5:137–145. PMID: 24322879.
Article30. Hashimoto K. Regulation of keratinocyte function by growth factors. J Dermatol Sci. 2000; 24(Suppl 1):S46–S50. PMID: 11137396.
Article31. Reichardt N, Duncan SH, Young P, Belenguer A, McWilliam Leitch C, Scott KP, et al. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 2014; 8:1323–1335. PMID: 24553467.
Article32. Thorburn AN, Macia L, Mackay CR. Diet, metabolites, and “western-lifestyle” inflammatory diseases. Immunity. 2014; 40:833–842. PMID: 24950203.
Article33. Kaikiri H, Miyamoto J, Kawakami T, Park SB, Kitamura N, Kishino S, et al. Supplemental feeding of a gut microbial metabolite of linoleic acid, 10-hydroxy-cis-12-octadecenoic acid, alleviates spontaneous atopic dermatitis and modulates intestinal microbiota in NC/nga mice. Int J Food Sci Nutr. 2017; 68:941–951. PMID: 28438083.
Article34. Matsumoto M, Ebata T, Hirooka J, Hosoya R, Inoue N, Itami S, et al. Antipruritic effects of the probiotic strain LKM512 in adults with atopic dermatitis. Ann Allergy Asthma Immunol. 2014; 113:209–216.e7. PMID: 24893766.
Article35. Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med. 2016; 22:1187–1191. PMID: 27618652.
Article36. Checa A, Holm T, Sjödin MO, Reinke SN, Alm J, Scheynius A, et al. Lipid mediator profile in vernix caseosa reflects skin barrier development. Sci Rep. 2015; 5:15740. PMID: 26521946.
Article37. Jin UH, Lee SO, Sridharan G, Lee K, Davidson LA, Jayaraman A, et al. Microbiome-derived tryptophan metabolites and their aryl hydrocarbon receptor-dependent agonist and antagonist activities. Mol Pharmacol. 2014; 85:777–788. PMID: 24563545.
Article38. Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012; 13:701–712. PMID: 22968153.
Article39. Yokoyama S, Hiramoto K, Koyama M, Ooi K. Impairment of skin barrier function via cholinergic signal transduction in a dextran sulphate sodium-induced colitis mouse model. Exp Dermatol. 2015; 24:779–784. PMID: 26014805.
Article40. Akiyama T, Iodi Carstens M, Carstens E. Transmitters and pathways mediating inhibition of spinal itch-signaling neurons by scratching and other counterstimuli. PLoS One. 2011; 6:e22665. PMID: 21818363.
Article41. Lee HJ, Park MK, Kim SY, Park Choo HY, Lee AY, Lee CH. Serotonin induces melanogenesis via serotonin receptor 2A. Br J Dermatol. 2011; 165:1344–1348. PMID: 21711335.
Article42. O'Neill CA, Monteleone G, McLaughlin JT, Paus R. The gut-skin axis in health and disease: A paradigm with therapeutic implications. BioEssays. 2016; 38:1167–1176. PMID: 27554239.43. Eyerich K, Novak N. Immunology of atopic eczema: overcoming the Th1/Th2 paradigm. Allergy. 2013; 68:974–982. PMID: 23889510.
Article44. Smith FJ, Irvine AD, Terron-Kwiatkowski A, Sandilands A, Campbell LE, Zhao Y, et al. Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat Genet. 2006; 38:337–342. PMID: 16444271.
Article45. Seite S, Bieber T. Barrier function and microbiotic dysbiosis in atopic dermatitis. Clin Cosmet Investig Dermatol. 2015; 8:479–483.
Article46. Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009; 324:1190–1192. PMID: 19478181.
Article47. Kennedy EA, Connolly J, Hourihane JO, Fallon PG, McLean WH, Murray D, et al. Skin microbiome before development of atopic dermatitis: early colonization with commensal staphylococci at 2 months is associated with a lower risk of atopic dermatitis at 1 year. J Allergy Clin Immunol. 2017; 139:166–172. PMID: 27609659.48. Nakatsuji T, Chen TH, Narala S, Chun KA, Two AM, Yun T, et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci Transl Med. 2017; 9:eaah4680. PMID: 28228596.
Article49. Thomas CL, Fernández-Peñas P. The microbiome and atopic eczema: more than skin deep. Australas J Dermatol. 2017; 58:18–24. PMID: 26821151.
Article50. Jun SH, Lee JH, Kim SI, Choi CW, Park TI, Jung HR, et al. Staphylococcus aureus-derived membrane vesicles exacerbate skin inflammation in atopic dermatitis. Clin Exp Allergy. 2017; 47:85–96. PMID: 27910159.51. Oh J, Freeman AF, Park M, Sokolic R, Candotti F, Holland SM, et al. The altered landscape of the human skin microbiome in patients with primary immunodeficiencies. Genome Res. 2013; 23:2103–2114. PMID: 24170601.
Article52. Nakatsuji T, Chen TH, Two AM, Chun KA, Narala S, Geha RS, et al. Staphylococcus aureus exploits epidermal barrier defects in atopic dermatitis to trigger cytokine expression. J Invest Dermatol. 2016; 136:2192–2200. PMID: 27381887.53. Laborel-Préneron E, Bianchi P, Boralevi F, Lehours P, Fraysse F, Morice-Picard F, et al. Effects of the Staphylococcus aureus and Staphylococcus epidermidis secretomes isolated from the skin microbiota of atopic children on CD4+ T cell activation. PLoS One. 2015; 10:e0141067. PMID: 26510097.54. Martin H, Laborel-Préneron E, Fraysse F, Nguyen T, Schmitt AM, Redoulès D, et al. Aquaphilus dolomiae extract counteracts the effects of cutaneous S. aureus secretome isolated from atopic children on CD4+ T cell activation. Pharm Biol. 2016; 54:2782–2785. PMID: 27180655.55. Kim MH, Rho M, Choi JP, Choi HI, Park HK, Song WJ, et al. A Metagenomic Analysis Provides a Culture-Independent Pathogen Detection for Atopic Dermatitis. Allergy Asthma Immunol Res. 2017; 9:453–461. PMID: 28677360.
Article56. Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012; 22:850–859. PMID: 22310478.
Article57. Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, et al. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature. 2010; 465:346–349. PMID: 20485435.58. Kobayashi T, Glatz M, Horiuchi K, Kawasaki H, Akiyama H, Kaplan DH, et al. Dysbiosis and Staphylococcus aureus colonization drives inflammation in atopic dermatitis. Immunity. 2015; 42:756–766. PMID: 25902485.59. Scharschmidt TC, Vasquez KS, Truong HA, Gearty SV, Pauli ML, Nosbaum A, et al. A wave of regulatory T cells into neonatal skin mediates tolerance to commensal microbes. Immunity. 2015; 43:1011–1021. PMID: 26588783.
Article60. Oh J, Byrd AL, Deming C, Conlan S, Kong HH, Segre JA. NISC Comparative Sequencing Program. Biogeography and individuality shape function in the human skin metagenome. Nature. 2014; 514:59–64. PMID: 25279917.
Article61. Chng KR, Tay AS, Li C, Ng AH, Wang J, Suri BK, et al. Whole metagenome profiling reveals skin microbiome-dependent susceptibility to atopic dermatitis flare. New Microbiol. 2016; 1:16106.
Article62. Lack G, Fox D, Northstone K, Golding J. Avon Longitudinal Study of Parents and Children Study Team. Factors associated with the development of peanut allergy in childhood. N Engl J Med. 2003; 348:977–985. PMID: 12637607.
Article63. Kelleher MM, Dunn-Galvin A, Gray C, Murray DM, Kiely M, Kenny L, et al. Skin barrier impairment at birth predicts food allergy at 2 years of age. J Allergy Clin Immunol. 2016; 137:1111–1116.e8. PMID: 26924469.64. Bartnikas LM, Gurish MF, Burton OT, Leisten S, Janssen E, Oettgen HC, et al. Epicutaneous sensitization results in IgE-dependent intestinal mast cell expansion and food-induced anaphylaxis. J Allergy Clin Immunol. 2013; 131:451–460.e1-6. PMID: 23374269.
Article65. Noti M, Kim BS, Siracusa MC, Rak GD, Kubo M, Moghaddam AE, et al. Exposure to food allergens through inflamed skin promotes intestinal food allergy through the thymic stromal lymphopoietinbasophil axis. J Allergy Clin Immunol. 2014; 133:1390–1399. 1399.e1–1399.e6. PMID: 24560412.
Article66. Galand C, Leyva-Castillo JM, Yoon J, Han A, Lee MS, McKenzie AN, et al. IL-33 promotes food anaphylaxis in epicutaneously sensitized mice by targeting mast cells. J Allergy Clin Immunol. 2016; 138:1356–1366. PMID: 27372570.
Article67. Wang Q, Du J, Zhu J, Yang X, Zhou B. Thymic stromal lymphopoietin signaling in CD4(+) T cells is required for TH2 memory. J Allergy Clin Immunol. 2015; 135:781–791.e3. PMID: 25441291.
Article68. Lee JB, Chen CY, Liu B, Mugge L, Angkasekwinai P, Facchinetti V, et al. IL-25 and CD4(+) TH2 cells enhance type 2 innate lymphoid cellderived IL-13 production, which promotes IgE-mediated experimental food allergy. J Allergy Clin Immunol. 2016; 137:1216–1225.e5. PMID: 26560039.
Article69. Chen CY, Lee JB, Liu B, Ohta S, Wang PY, Kartashov AV, et al. Induction of interleukin-9-producing mucosal mast cells promotes susceptibility to IgE-mediated experimental food allergy. Immunity. 2015; 43:788–802. PMID: 26410628.
Article70. Mondoulet L, Dioszeghy V, Puteaux E, Ligouis M, Dhelft V, Plaquet C, et al. Specific epicutaneous immunotherapy prevents sensitization to new allergens in a murine model. J Allergy Clin Immunol. 2015; 135:1546–1557.e4. PMID: 25583102.
Article71. Akbari O, DeKruyff RH, Umetsu DT. Pulmonary dendritic cells producing IL-10 mediate tolerance induced by respiratory exposure to antigen. Nat Immunol. 2001; 2:725–731. PMID: 11477409.
Article72. Watanabe S, Narisawa Y, Arase S, Okamatsu H, Ikenaga T, Tajiri Y, et al. Differences in fecal microflora between patients with atopic dermatitis and healthy control subjects. J Allergy Clin Immunol. 2003; 111:587–591. PMID: 12642841.
Article73. Mah KW, Björkstén B, Lee BW, van Bever HP, Shek LP, Tan TN, et al. Distinct pattern of commensal gut microbiota in toddlers with eczema. Int Arch Allergy Immunol. 2006; 140:157–163. PMID: 16601353.
Article74. Penders J, Thijs C, van den Brandt PA, Kummeling I, Snijders B, Stelma F, et al. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study. Gut. 2007; 56:661–667. PMID: 17047098.
Article75. van Nimwegen FA, Penders J, Stobberingh EE, Postma DS, Koppelman GH, Kerkhof M, et al. Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. J Allergy Clin Immunol. 2011; 128:948–955.e1-3. PMID: 21872915.
Article76. Penders J, Gerhold K, Stobberingh EE, Thijs C, Zimmermann K, Lau S, et al. Establishment of the intestinal microbiota and its role for atopic dermatitis in early childhood. J Allergy Clin Immunol. 2013; 132:601–607.e8. PMID: 23900058.
Article77. Shi B, Bangayan NJ, Curd E, Taylor PA, Gallo RL, Leung DY, et al. The skin microbiome is different in pediatric versus adult atopic dermatitis. J Allergy Clin Immunol. 2016; 138:1233–1236. PMID: 27474122.