Ann Lab Med.
2019 Jan;39(1):43-49. 10.3343/alm.2019.39.1.43.
Shorter Incubation Times for Detecting Multi-drug Resistant Bacteria in Patient Samples: Defining Early Imaging Time Points Using Growth Kinetics and Total Laboratory Automation
- Affiliations
-
- 1Department for Infectious Diseases, Microbiology and Hygiene, University Hospital of Heidelberg, Germany. irene.burckhardt@med.uni-heidelberg.de
- KMID: 2420270
- DOI: http://doi.org/10.3343/alm.2019.39.1.43
Abstract
- BACKGROUND
The transition from manual processing of patient samples to automated workflows in medical microbiology is challenging. Although automation enables microbiologists to evaluate all samples following the same incubation period, the essential incubation times have yet to be determined. We defined essential incubation times for detecting methicillin-resistant Staphylococcus aureus (MRSA), multi-drug resistant gram-negative bacteria (MDRGN), and vancomycin-resistant enterococci (VRE).
METHODS
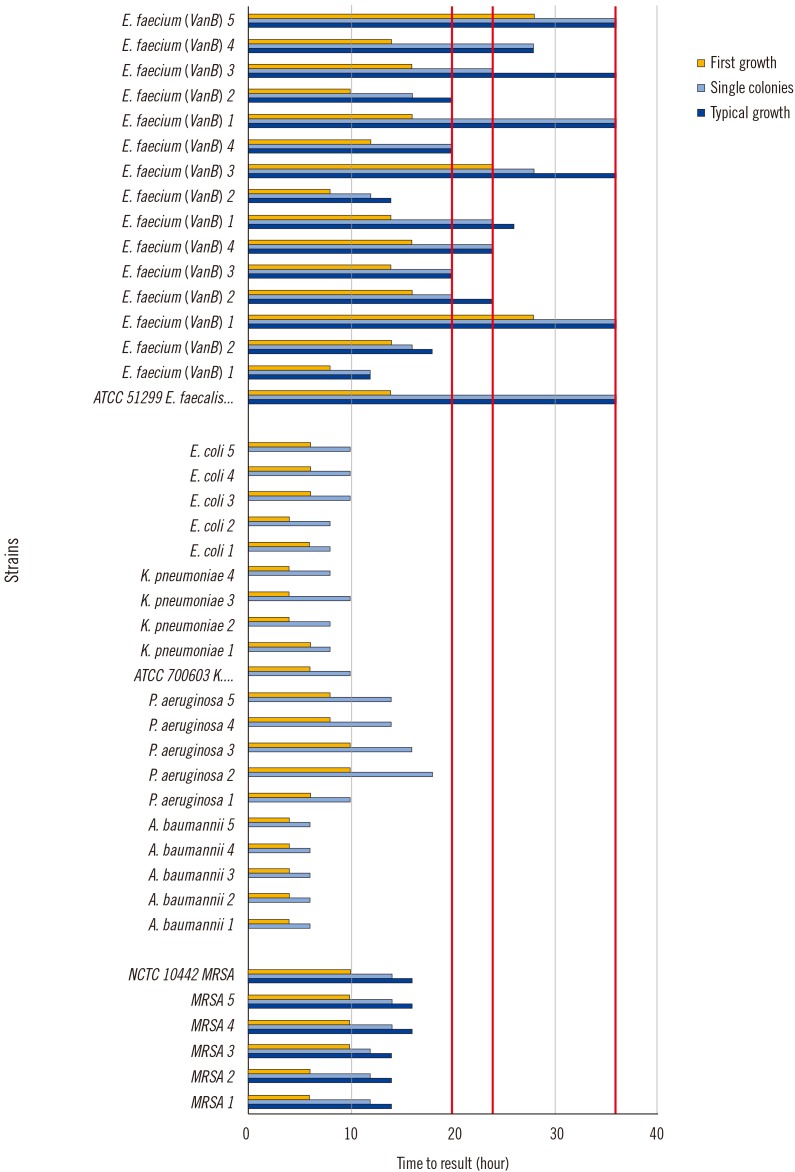
We monitored the growth kinetics of MRSA, MDRGN, and VRE between two and 48 hours on chromogenic media to establish the time points of first growth, single colony appearance, and typical morphology for 102, 104, 106, and 108 colony forming units/mL. Subsequently, we imaged plates inoculated with 778 patient samples after 20, 24, and 36 hours.
RESULTS
The first growth, single colony appearance, and typical morphology time points were inoculum-dependent. First growth appeared after 6-18 hours, 4-18 hours, and 8-48 hours for MRSA, MDRGN, and VRE, respectively, and single colonies appeared at 12-18 hours, 6-20 hours, and 12-48 hours, respectively. Typical morphology was visible at 14-22 hours and 12-48 hours for MRSA and VRE, but was not determined for MDRGN. By examining patient samples, ≥98% of MRSA and MDRGN were visible 20 hours after the start of incubation. Following 24 hours of incubation, only 79.5% of VRE were clearly visible on the respective plates.
CONCLUSIONS
An incubation time of 20 hours is sufficient for detecting MRSA and MDRGN. VRE growth is much slower and requires additional imaging after 36 hours.
Keyword
MeSH Terms
Figure
Reference
-
1. Rubinovitch B, Pittet D. Screening for methicillin-resistant Staphylococcus aureus in the endemic hospital: what have we learned? J Hosp Infect. 2001; 47:9–18. PMID: 11161895.2. Hagen RM, Seegmüller I, Navai J, Kappstein I, Lehn N, Miethke T. Development of a real-time PCR assay for rapid identification of methicillin-resistant Staphylococcus aureus from clinical samples. Int J Med Microbiol. 2005; 295:77–86. PMID: 15969468.3. Dalpke AH, Hofko M, Zimmermann S. Development of a real-time PCR protocol requiring minimal handling for detection of vancomycin-resistant enterococci with the fully automated BD Max system. J Clin Microbiol. 2016; 54:2321–2329. PMID: 27358466.4. Blanc DS, Basset P, Nahimana-Tessemo I, Jaton K, Greub G, Zanetti G. High proportion of wrongly identified methicillin-resistant Staphylococcus aureus carriers by use of a rapid commercial PCR assay due to presence of staphylococcal cassette chromosome element lacking the mecA gene. J Clin Microbiol. 2011; 49:722–724. PMID: 21159945.5. van den Bijllaardt W, Buiting AG, Mouton JW, Muller AE. Shortening the incubation time for antimicrobial susceptibility testing by disk diffusion for Enterobacteriaceae: how short can it be and are the results accurate? Int J Antimicrob Agents. 2017; 49:631–637. PMID: 28263895.6. Fröding I, Vondracek M, Giske CG. Rapid EUCAST disc diffusion testing of MDR Escherichia coli and Klebsiella pneumoniae: inhibition zones for extended-spectrum cephalosporins can be reliably read after 6 h of incubation. J Antimicrob Chemother. 2017; 72:1094–1102. PMID: 27999046.7. Hombach M, Jetter M, Blöchliger N, Kolesnik-Goldmann N, Böttger EC. Fully automated disc diffusion for rapid antibiotic susceptibility test results: a proof-of-principle study. J Antimicrob Chemother. 2017; 72:1659–1668. PMID: 28333189.8. Burckhardt I, Panitz J, Burckhardt F, Zimmermann S. Identification of Streptococcus pneumoniae: development of a standardized protocol for optochin susceptibility testing using total lab automation. Biomed Res Int. 2017; 2017:4174168. PMID: 28401155.9. Hygiene measures for infection or colonization with multidrug-resistant gram-negative bacilli. Commission recommendation for hospital hygiene and infection prevention (KRINKO) at the Robert Koch Institute (RKI). Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2012; 55:1311–1354. PMID: 23011096.10. Mattner F, Bange FC, Meyer E, Seifert H, Wichelhaus TA, Chaberny IF. Preventing the spread of multidrug-resistant gram-negative pathogens: recommendations of an expert panel of the German Society for Hygiene and Microbiology. Dtsch Arztebl Int. 2012; 109:39–45. PMID: 22334820.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Performance of Kiestra Total Laboratory Automation Combined with MS in Clinical Microbiology Practice
- Quantitative Analysis of Weissella cibaria against Periodontopathic Bacteria by Real-time PCR
- Modified CHROMagar Acinetobacter Medium for Direct Detection of Multidrug-Resistant Acinetobacter Strains in Nasal and Rectal Swab Samples
- Effectiveness of Nanocrystalline Silver(Acticoat(R) Dressing at Wound Infected by Multidrug Resistant Bacteria
- Comparison of Sensitivity of Tests for Detecting Bacterial Contamination in Platelet Concentrates