Intest Res.
2018 Apr;16(2):246-254. 10.5217/ir.2018.16.2.246.
Characterization of the fecal microbiota differs between age groups in Koreans
- Affiliations
-
- 1Probioticslab R&D Institute, Bioeleven Co., Seoul, Korea. jinkim@bio11.co.kr
- 2Department of Pediatric, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea.
- 3Department of Internal Medicine, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea.
- KMID: 2417676
- DOI: http://doi.org/10.5217/ir.2018.16.2.246
Abstract
- BACKGROUND/AIMS
Tens of trillions of microorganisms constitute the gut microbiota of the human body. The microbiota plays a critical role in maintaining host immunity and metabolism. Analyses of the gut microbial composition in Korea are limited to a few studies consisting of small sample sizes. To investigate the gut microbial community in a large sample of healthy Koreans, we analyzed the 16S ribosomal RNA of 4 representative bacterial genera Lactobacillus, Bifidobacterium, Bacteroides, and Clostridium.
METHODS
A total of 378 DNA samples extracted from 164 infants and 214 adults were analyzed using quantitative real-time polymerase chain reaction.
RESULTS
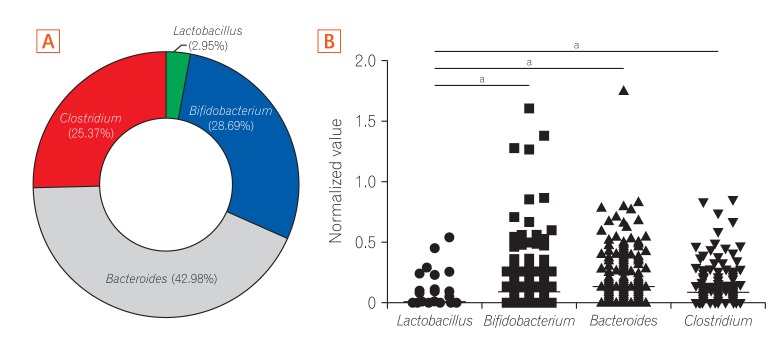
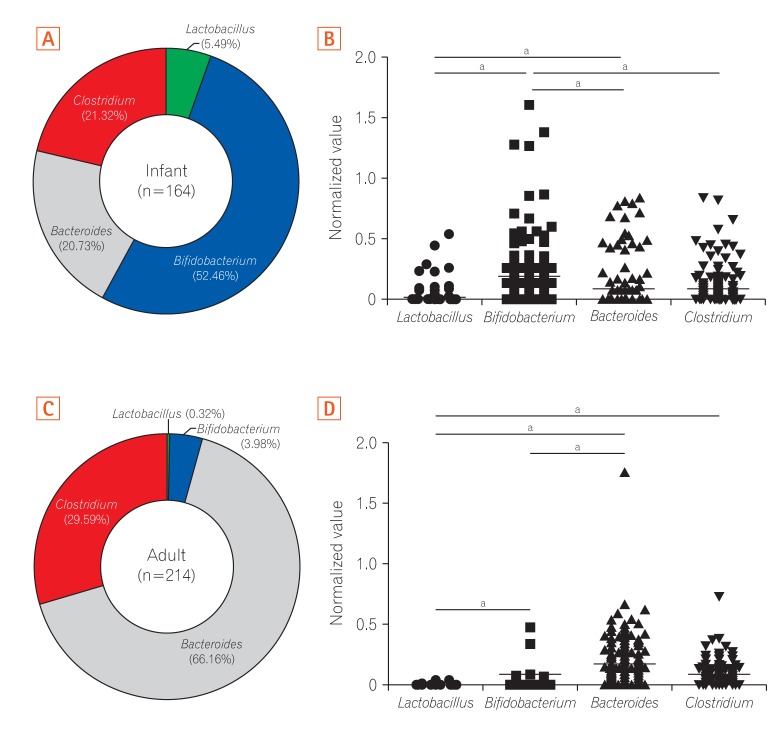
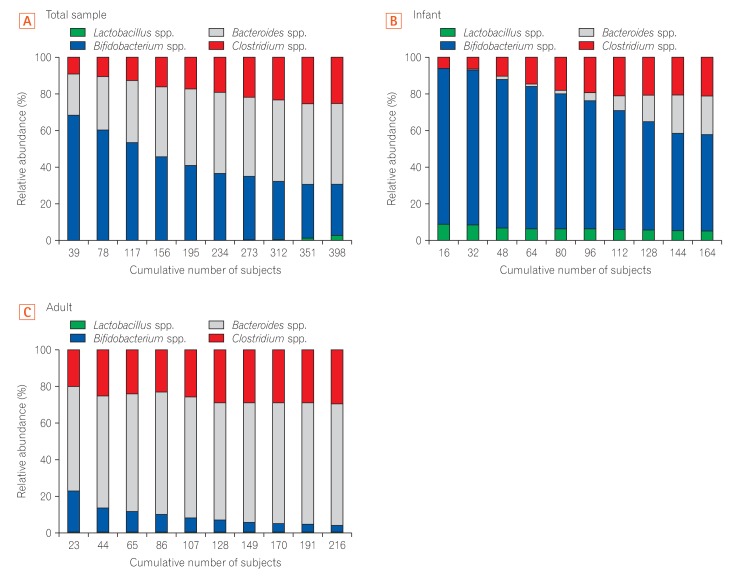
Analysis of 16S ribosomal RNA of 4 representative bacterial genera Lactobacillus, Bifidobacterium, Bacteroides, and Clostridium showed that the gut microbiota in infants had higher relative abundances of Bifidobacterium and Lactobacillus than that in adults, which was dominated by Bacteroides and Clostridium.
CONCLUSIONS
To the best of our knowledge, this was the first study evaluating the distinct characteristics of the microbial community of Korean infants and adults. The differences between the 2 populations suggest that external factors such as age, diet, and the environment are important contributing factors to the change in gut microbial composition during development.
Keyword
MeSH Terms
Figure
Cited by 1 articles
-
Characteristics of Faecal Microbiota in Korean Patients with Clostridioides difficile-associated Diarrhea
Yong Duk Jeon, Hea Won Ann, Woon Ji Lee, Jun Hyoung Kim, Hye Seong, Jung Ho Kim, Jin Young Ahn, Su Jin Jeong, Nam Su Ku, Joon Sup Yeom, Dongeun Yong, Kyungwon Lee, Jun Yong Choi
Infect Chemother. 2019;51(4):365-375. doi: 10.3947/ic.2019.51.4.365.
Reference
-
1. Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005; 308:1635–1638. PMID: 15831718.2. Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010; 107:11971–11975. PMID: 20566857.3. Mitsuoka T. Bifidobacteria and their role in human health. J Ind Microbiol Biotechnol. 1990; 6:263–267.4. Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007; 5:e177. DOI: 10.1371/journal.pbio.0050177. PMID: 17594176.5. Suzuki TA, Worobey M. Geographical variation of human gut microbial composition. Biol Lett. 2014; 10:20131037. DOI: 10.1098/rsbl.2013.1037. PMID: 24522631.6. David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014; 505:559–563. PMID: 24336217.7. Nicholson JK, Holmes E, Kinross J, et al. Host-gut microbiota metabolic interactions. Science. 2012; 336:1262–1267. PMID: 22674330.
Article8. Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009; 457:480–484. PMID: 19043404.
Article9. de la Cochetiere MF, Piloquet H, des Robert C, Darmaun D, Galmiche JP, Roze JC. Early intestinal bacterial colonization and necrotizing enterocolitis in premature infants: the putative role of Clostridium. Pediatr Res. 2004; 56:366–370. PMID: 15201403.
Article10. Fell JM. Neonatal inflammatory intestinal diseases: necrotising enterocolitis and allergic colitis. Early Hum Dev. 2005; 81:117–122. PMID: 15707723.
Article11. Finegold SM. Therapy and epidemiology of autism: clostridial spores as key elements. Med Hypotheses. 2008; 70:508–511. PMID: 17904761.12. O'Keefe SJ. Nutrition and colonic health: the critical role of the microbiota. Curr Opin Gastroenterol. 2008; 24:51–58. PMID: 18043233.13. Marchesi JR, Holmes E, Khan F, et al. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J Proteome Res. 2007; 6:546–551. PMID: 17269711.
Article14. Penders J, Vink C, Driessen C, London N, Thijs C, Stobberingh EE. Quantification of Bifidobacterium spp., Escherichia coli and Clostridium difficile in faecal samples of breast-fed and formula-fed infants by real-time PCR. FEMS Microbiol Lett. 2005; 243:141–147. PMID: 15668012.
Article15. Giaffer MH, Holdsworth CD, Duerden BI. The assessment of faecal flora in patients with inflammatory bowel disease by a simplified bacteriological technique. J Med Microbiol. 1991; 35:238–243. PMID: 1941994.
Article16. Madsen KL, Doyle JS, Jewell LD, Tavernini MM, Fedorak RN. Lactobacillus species prevents colitis in interleukin 10 gene-deficient mice. Gastroenterology. 1999; 116:1107–1114. PMID: 10220502.
Article17. Mencarelli A, Distrutti E, Renga B, et al. Probiotics modulate intestinal expression of nuclear receptor and provide counter-regulatory signals to inflammation-driven adipose tissue activation. PLoS One. 2011; 6:e22978. DOI: 10.1371/journal.pone.0022978. PMID: 21829567.
Article18. Kerckhoffs AP, Samsom M, van der Rest ME, et al. Lower Bifidobacteria counts in both duodenal mucosa-associated and fecal microbiota in irritable bowel syndrome patients. World J Gastroenterol. 2009; 15:2887–2892. PMID: 19533811.
Article19. Cox LM, Yamanishi S, Sohn J, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014; 158:705–721. PMID: 25126780.
Article20. Kostic AD, Gevers D, Siljander H, et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe. 2015; 17:260–273. PMID: 25662751.
Article21. Suau A, Bonnet R, Sutren M, et al. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol. 1999; 65:4799–4807. PMID: 10543789.
Article22. Kim SE, Choi SC, Park KS, et al. Change of fecal flora and effectiveness of the short-term VSL#3 probiotic treatment in patients with functional constipation. J Neurogastroenterol Motil. 2015; 21:111–120. PMID: 25537674.
Article23. Kwok LY, Zhang J, Guo Z, et al. Characterization of fecal microbiota across seven Chinese ethnic groups by quantitative polymerase chain reaction. PLoS One. 2014; 9:e93631. DOI: 10.1371/journal.pone.0093631. PMID: 24699404.
Article24. Carroll IM, Chang YH, Park J, Sartor RB, Ringel Y. Luminal and mucosal-associated intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Gut Pathog. 2010; 2:19. DOI: 10.1186/1757-4749-2-19. PMID: 21143915.
Article25. Wang Y, Ametaj BN, Ambrose DJ, Gänzle MG. Characterisation of the bacterial microbiota of the vagina of dairy cows and isolation of pediocin-producing Pediococcus acidilactici. BMC Microbiol. 2013; 13:19. DOI: 10.1186/1471-2180-13-19. PMID: 23356904.
Article26. Matsuki T, Watanabe K, Fujimoto J, et al. Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Appl Environ Microbiol. 2002; 68:5445–5451. PMID: 12406736.
Article27. Rinttilä T, Kassinen A, Malinen E, Krogius L, Palva A. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J Appl Microbiol. 2004; 97:1166–1177. PMID: 15546407.
Article28. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008; 3:1101–1108. PMID: 18546601.
Article29. Sjögren YM, Jenmalm MC, Böttcher MF, Björkstén B, Sverremark-Ekström E. Altered early infant gut microbiota in children developing allergy up to 5 years of age. Clin Exp Allergy. 2009; 39:518–526. PMID: 19220322.
Article30. Hopkins MJ, Sharp R, Macfarlane GT. Age and disease related changes in intestinal bacterial populations assessed by cell culture, 16S rRNA abundance, and community cellular fatty acid profiles. Gut. 2001; 48:198–205. PMID: 11156640.
Article31. Tap J, Mondot S, Levenez F, et al. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol. 2009; 11:2574–2584. PMID: 19601958.32. Kurokawa K, Itoh T, Kuwahara T, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007; 14:169–181. PMID: 17916580.33. Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008; 6:e280. DOI: 10.1371/journal.pbio.0060280. PMID: 19018661.34. Gill SR, Pop M, Deboy RT, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006; 312:1355–1359. PMID: 16741115.35. Bélanger SD, Boissinot M, Clairoux N, Picard FJ, Bergeron MG. Rapid detection of Clostridium difficile in feces by real-time PCR. J Clin Microbiol. 2003; 41:730–734. PMID: 12574274.36. Huijsdens XW, Linskens RK, Mak M, Meuwissen SG, Vandenbroucke-Grauls CM, Savelkoul PH. Quantification of bacteria adherent to gastrointestinal mucosa by real-time PCR. J Clin Microbiol. 2002; 40:4423–4427. PMID: 12454130.
Article37. Swidsinski A, Weber J, Loening-Baucke V, Hale LP, Lochs H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol. 2005; 43:3380–3389. PMID: 16000463.
Article38. Kassinen A, Krogius-Kurikka L, Mäkivuokko H, et al. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology. 2007; 133:24–33. PMID: 17631127.
Article39. Malinen E, Rinttilä T, Kajander K, et al. Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy controls with real-time PCR. Am J Gastroenterol. 2005; 100:373–382. PMID: 15667495.
Article40. Distrutti E, O'Reilly JA, McDonald C, et al. Modulation of intestinal microbiota by the probiotic VSL#3 resets brain gene expression and ameliorates the age-related deficit in LTP. PLoS One. 2014; 9:e106503. DOI: 10.1371/journal.pone.0106503. PMID: 25202975.
Article41. Nam YD, Jung MJ, Roh SW, Kim MS, Bae JW. Comparative analysis of Korean human gut microbiota by barcoded pyrosequencing. PLoS One. 2011; 6:e22109. DOI: 10.1371/journal.pone.0022109. PMID: 21829445.
Article42. Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011; 473:174–180. PMID: 21508958.
Article43. Turroni F, Peano C, Pass DA, et al. Diversity of bifidobacteria within the infant gut microbiota. PLoS One. 2012; 7:e36957. DOI: 10.1371/journal.pone.0036957. PMID: 22606315.
Article44. Lozupone CA, Stombaugh J, Gonzalez A, et al. Meta-analyses of studies of the human microbiota. Genome Res. 2013; 23:1704–1714. PMID: 23861384.
Article45. Jakobsson HE, Jernberg C, Andersson AF, Sjölund-Karlsson M, Jansson JK, Engstrand L. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One. 2010; 5:e9836. DOI: 10.1371/journal.pone.0009836. PMID: 20352091.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Fecal Microbiota Transplantation and the Brain Microbiota in Neurological Diseases
- Clinical Usefulness of Fecal Microbiota Transplantation
- Refractory Clostridium difficile Infection Cured With Fecal Microbiota Transplantation in Vancomycin-Resistant Enterococcus Colonized Patient
- Is there a potential role of fecal microbiota transplantation in the treatment of inflammatory bowel disease?
- Excrement Strikes Back: The Dark Side of Fecal Microbiota Transplantation