J Vet Sci.
2017 Jun;18(2):217-227. 10.4142/jvs.2017.18.2.217.
Bovine adenovirus type 3 virions cannot be rescued in vivo after full-length viral genome transfection in the absence of detectable polypeptide IX
- Affiliations
-
- 1College of Veterinary Medicine, North-west A&F University, Yangling 712100, China. duenqi227@126.com
- 2Chinese Institute of Veterinary Drug Controls, Beijing 100000, China.
- 3VIDO-InteVac, School of Public Health, University of Saskatchewan Saskatoon, Saskatchewan S4S 6A6, Canada. duenqi227@126.com
- 4Vaccinology & Immunotherapeutics Program, School of Public Health, University of Saskatchewan Saskatoon, Saskatchewan S4S 6A6, Canada.
- KMID: 2412575
- DOI: http://doi.org/10.4142/jvs.2017.18.2.217
Abstract
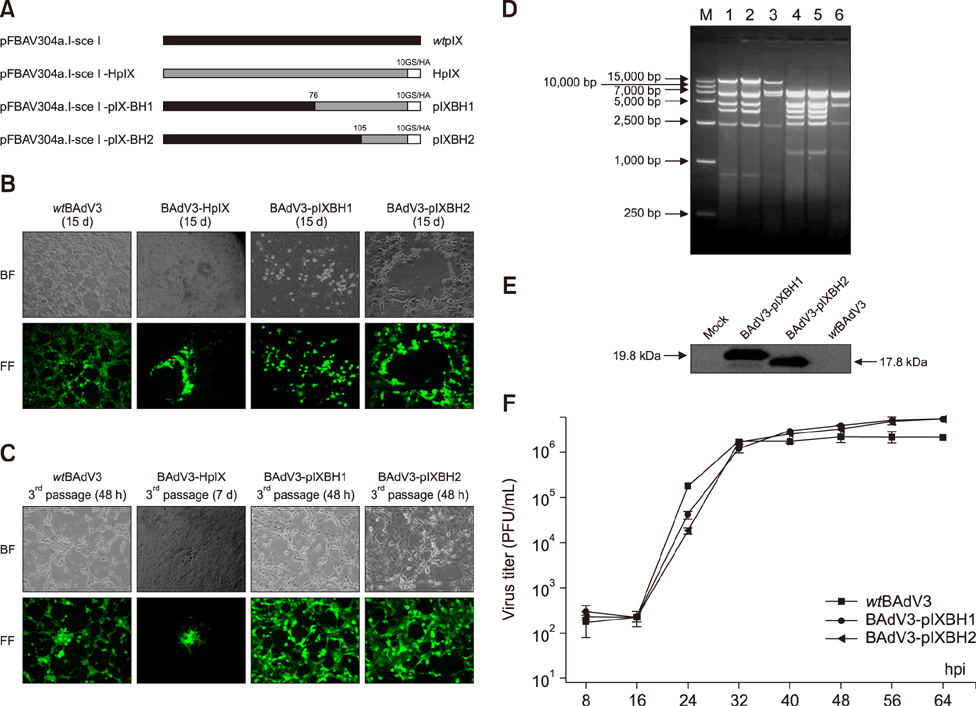
- Bovine adenovirus type 3 (BAdV3) is being used in the development of potential vehicles for gene therapy and vectored vaccine. To that end, a more comprehensive description of BAdV3 biology is essential. In this study, we focused on the role of pIX in BAdV3 virion rescue after full-length BAdV3 genome transfection. Initially, pIX deletion or initiation codon mutation abolished the production of progeny virions, which suggested that pIX was essential for the rescue of BAdV3 containing a full-length genome. Moreover, through transfection of a panel of pIX mutant BAdV3 genomes, we observed that the conserved N-terminus and the putative leucine zipper element (PLZP) were essential for virion rescue, whereas the C-terminus following the coiled-coil domain was non-essential. In addition, swap of the PLZP element and its following region of BAdV3 pIX to corresponding domains of human adenovirus type 5 (HAdV5) did not affect virion production, whereas swap of the entire pIX abolished production of progeny virions. We suggest that failure of the full-length BAdV3 pIX swap might be due to species specificity of its N-terminus region before the PLZP element.
MeSH Terms
Figure
Reference
-
1. Bangari DS, Shukla S, Mittal SK. Comparative transduction efficiencies of human and nonhuman adenoviral vectors in human, murine, bovine, and porcine cells in culture. Biochem Biophys Res Commun. 2005; 327:960–966.
Article2. Blanchette P, Wimmer P, Dallaire F, Cheng CY, Branton PE. Aggresome formation by the adenoviral protein E1B55K is not conserved among adenovirus species and is not required for efficient degradation of nuclear substrates. J Virol. 2013; 87:4872–4881.
Article3. Cadiñanos J, Bradley A. A Generation of an inducible and optimized piggyBac transposon system. Nucleic Acids Res. 2007; 35:e87.4. Cheng CY, Gilson T, Wimmer P, Schreiner S, Ketner G, Dobner T, Branton PE, Blanchette P. Role of E1B55K in E4orf6/E1B55K E3 ligase complexes formed by different human adenovirus serotypes. J Virol. 2013; 87:6232–6245.
Article5. Colby WW, Shenk T. Adenovirus type 5 virions can be assembled in vivo in the absence of detectable polypeptide IX. J Virol. 1981; 39:977–980.
Article6. Du E, Tikoo SK. Efficient replication and generation of recombinant bovine adenovirus-3 in nonbovine cotton rat lung cells expressing I-SceI endonuclease. J Gene Med. 2010; 12:840–847.
Article7. Fabry CM, Rosa-Calatrava M, Moriscot C, Ruigrok RW, Boulanger P, Schoehn G. The C-terminal domains of adenovirus serotype 5 protein IX assemble into an antiparallel structure on the facets of the capsid. J Virol. 2009; 83:1135–1139.
Article8. Ferreira TB, Alves PM, Aunins JG, Carrondo MJ. Use of adenoviral vectors as veterinary vaccines. Gene Ther. 2005; 12(Suppl 1):S73–S83.
Article9. Furcinitti PS, van Oostrum J, Burnett RM. Adenovirus polypeptide IX revealed as capsid cement by difference images from electron microscopy and crystallography. EMBO J. 1989; 8:3563–3570.
Article10. Ghosh-Choudhury G, Haj-Ahmad Y, Graham FL. Protein IX, a minor component of the human adenovirus capsid, is essential for the packaging of full length genomes. EMBO J. 1987; 6:1733–1739.
Article11. Kunec D, Hanson LA, van Haren S, Nieuwenhuizen IF, Burgess SC. An overlapping bacterial artificial chromosome system that generates vectorless progeny for channel catfish herpesvirus. J Virol. 2008; 82:3872–3881.
Article12. Liu H, Jin L, Koh SBS, Atanasov I, Schein S, Wu L, Zhou ZH. Atomic structure of human adenovirus by cryoEM reveals interactions among protein networks. Science. 2010; 329:1038–1043.
Article13. Lutz P, Rosa-Calatrava M, Kedinger C. The product of the adenovirus intermediate gene IX is a transcriptional activator. J Virol. 1997; 71:5102–5109.
Article14. Parks RJ. Adenovirus protein IX: a new look at an old protein. Mol Ther. 2005; 11:19–25.
Article15. Parks RJ, Graham FL. A helper-dependent system for adenovirus vector production helps define a lower limit for efficient DNA packing. J Virol. 1997; 71:3293–3298.
Article16. Reddy PS, Chen Y, Idamakanti N, Pyne C, Babiuk LA, Tikoo SK. Characterization of early region 1 and pIX of bovine adenovirus-3. Virology. 1999; 253:299–308.
Article17. Reddy PS, Idamakanti N, Zakhartchouk AN, Baxi MK, Lee JB, Pyne C, Babiuk LA, Tikoo SK. Nucleotide sequence, genome organization, and transcription map of bovine adenovirus type 3. J Virol. 1998; 72:1394–1402.
Article18. Rosa-Calatrava M, Grave L, Puvion-Dutilleul F, Chatton B, Kedinger C. Functional analysis of adenovirus protein IX identifies domains involved in capsid stability, transcriptional activity, and nuclear reorganization. J Virol. 2001; 75:7131–7141.
Article19. Rosa-Calatrava M, Puvion-Dutilleul F, Lutz P, Dreyer D, de Thé H, Chatton B, Kedinger C. Adenovirus protein IX sequesters host-cell promyelocytic leukaemia protein and contributes to efficient viral proliferation. EMBO Rep. 2003; 4:969–975.
Article20. Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010; 5:725–738.
Article21. Sargent KL, Meulenbroek RA, Parks RJ. Activation of adenoviral gene expression by protein IX is not required for efficient virus replication. J Virol. 2004; 78:5032–5037.
Article22. Souquere-Besse S, Pichard E, Filhol O, Legrand V, Rosa-Calatrava M, Hovanessian AG, Cochet C, Puvion-Dutilleul F. Adenovirus infection targets the cellular protein kinase CK2 and RNA-activated protein kinase (PKR) into viral inclusions of the cell nucleus. Microsc Res Tech. 2002; 56:465–478.
Article23. Stewart PL, Fuller SD, Burnett RM. Difference imaging of adenovirus: bridging the resolution gap between X-ray crystallography and electron microscopy. EMBO J. 1993; 12:2589–2599.
Article24. Strunze S, Engelke MF, Wang IH, Puntener D, Boucke K, Schleich S, Way M, Schoenenberger P, Burckhardt CJ, Greber UF. Kinesin-1-mediated capsid disassembly and disruption of the nuclear pore complex promote virus infection. Cell Host Microbe. 2011; 10:210–223.
Article25. van Oostrum J, Burnett RM. Molecular composition of the adenovirus type 2 virion. J Virol. 1985; 56:439–448.
Article26. Vellinga J, van den Wollenberg DJ, van der Heijdt S, Rabelink MJ, Hoeben RC. The coiled-coil domain of the adenovirus type 5 protein IX is dispensable for capsid incorporation and thermostability. J Virol. 2005; 79:3206–3210.
Article27. Zakhartchouk A, Connors W, van Kessel A, Tikoo SK. Bovine adenovirus type 3 containing heterologous protein in the C-terminus of minor capsid protein IX. Virology. 2004; 320:291–300.
Article28. Zakhartchouk AN, Pyne C, Mutwiri GK, Papp Z, Baca-Estrada ME, Griebel P, Babiuk LA, Tikoo SK. Mucosal immunization of calves with recombinant bovine adenovirus-3: induction of protective immunity to bovine herpesvirus-1. J Gen Virol. 1999; 80:1263–1269.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Expression and Packaging of a Human Endogenous Retrovirus-K Genomic DNA Clone
- Construction of Recombinant p53 Adenovirus ( Ad5CMVp53 ) for Cervical Cancer Gene Therapy
- Construction of Recombinant Adeno-Associated Virus Vector (AAVCMVp53) for Human Cervical Cancer Gene Therapy
- Biochemical properties of full-length hepatitis C virus RNA-dependent RNA polymerase expressed in insect cells
- Enhancement of Adenovirus Type 12 Transformation by N-Methyl-N-Nitro-N-Nitrosoguanidine