J Vet Sci.
2018 May;19(3):406-415. 10.4142/jvs.2018.19.3.406.
Identification, sequence analysis, and infectivity of H9N2 avian influenza viruses isolated from geese
- Affiliations
-
- 1College of Veterinary Medicine, Yangzhou University, Yangzhou 225009, China. hyshi@yzu.edu.cn
- 2Jiangsu Co-innovation Center for the Prevention and Control of Important Animal Infectious Diseases and Zoonoses, Yangzhou 225009, China.
- 3Sinopharm Yangzhou VAC Biological Engineering Co., Ltd., Yangzhou 225009, China.
- 4Department of Infectious Diseases and Pathology, College of Veterinary Medicine, University of Florida, Gainesville, FL 32611, USA.
- KMID: 2412131
- DOI: http://doi.org/10.4142/jvs.2018.19.3.406
Abstract
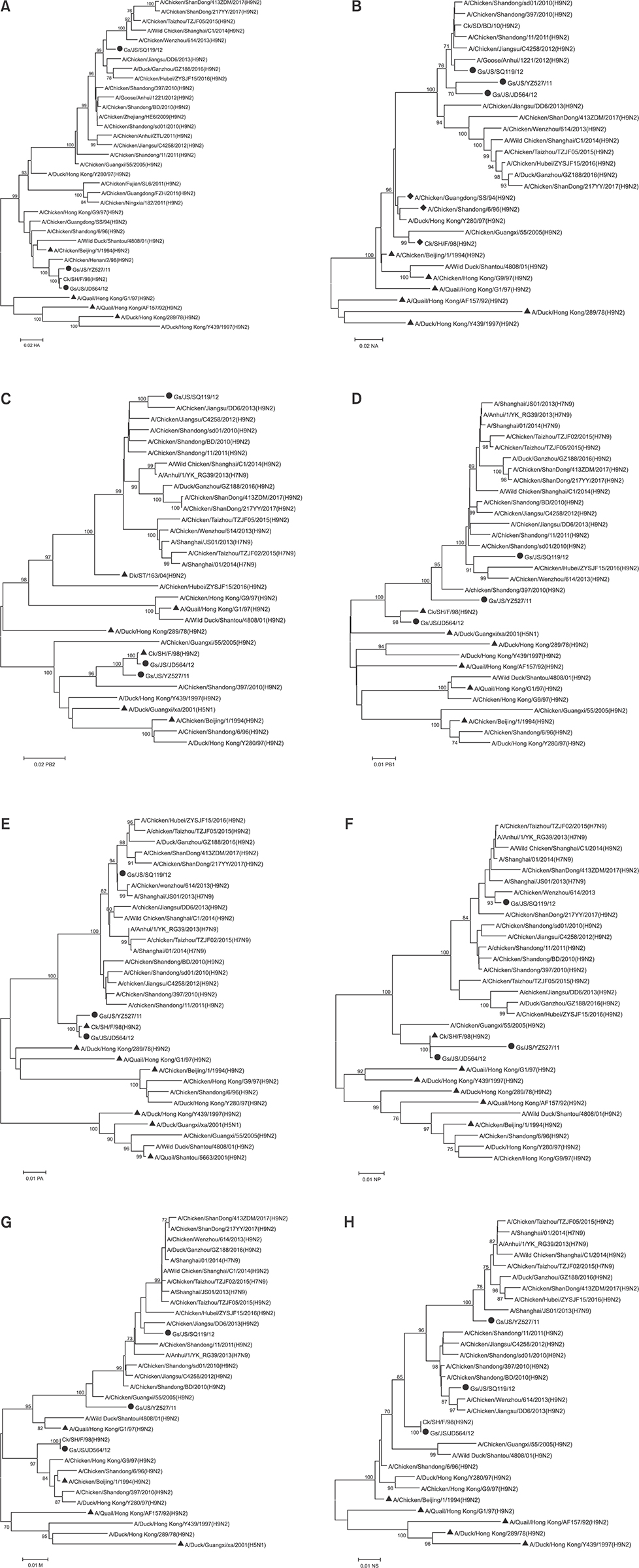
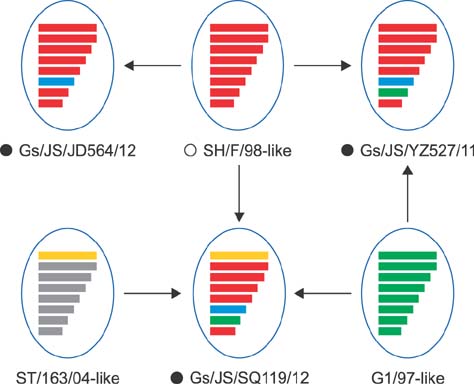
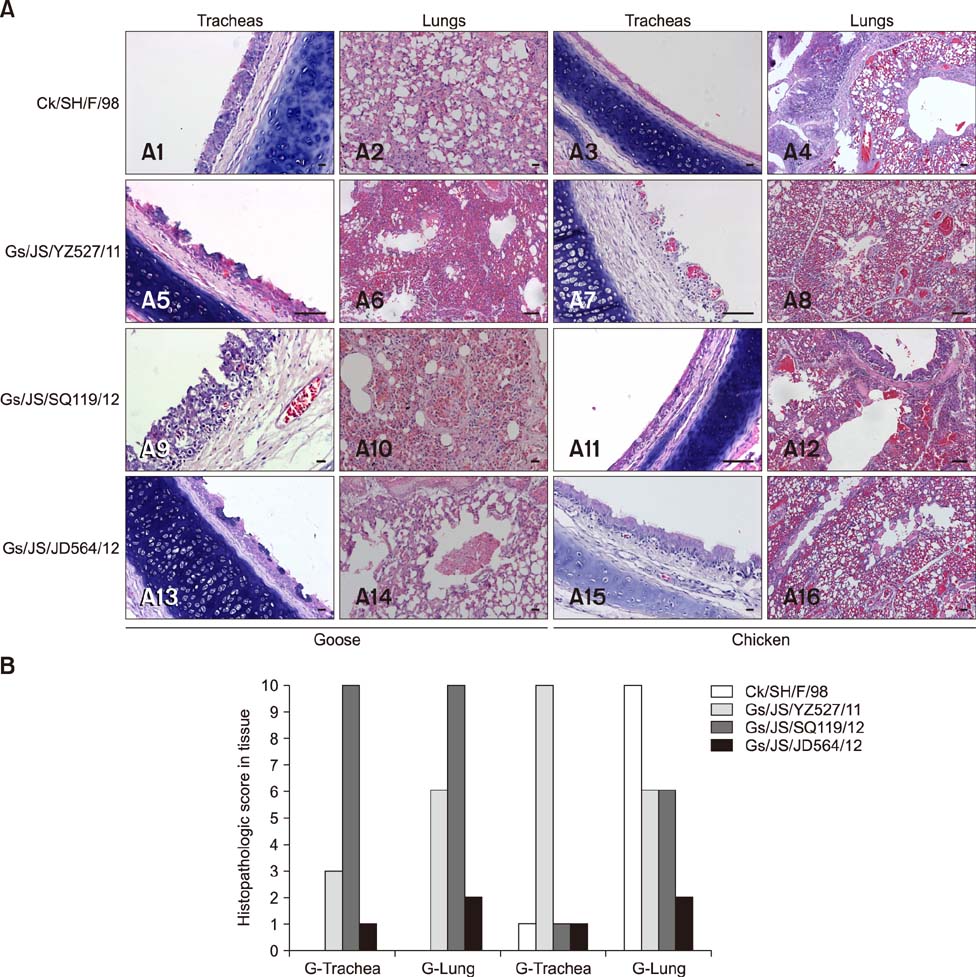
- The subtype H9N2 avian influenza virus greatly threatens the Chinese poultry industry, even with annual vaccination. Waterfowl can be asymptomatically infected with the H9N2 virus. In this study, three H9N2 virus strains, designated A/Goose/Jiangsu/YZ527/2011 (H9N2, Gs/JS/YZ527/11), A/Goose/Jiangsu/SQ119/2012 (H9N2, Gs/JS/SQ119/12), and A/Goose/Jiangsu/JD564/2012 (H9N2, Gs/JS/JD564/12), were isolated from domestic geese. Molecular characterization of the three isolates showed that the Gs/JS/YZ527/11 virus is a double-reassortant virus, combining genes of A/Quail/Hong Kong/G1/97 (H9N2, G1/97)-like and A/Chicken/Shanghai/F/98 (H9N2, F/98)-like; the Gs/JS/SQ119/12 virus is a triple-reassortant virus combining genes of G1/97-like, F/98-like, and A/Duck/Shantou/163/2004 (H9N2, ST/163/04)-like. The sequences of Gs/JS/JD564/12 share high homology with those of the F/98 virus, except for the neuraminidase gene, whereas the internal genes of Gs/JS/YZ527/11 and Gs/JS/SQ119/12 are closely related to those of the H7N9 viruses. An infectivity analysis of the three isolates showed that Gs/JS/SQ119/12 and Gs/JS/YZ527/11 replicated well, with seroconversion, in geese and chickens, the Gs/JS/JD564/12 did not infect well in geese or chickens, and the F/98 virus only infected chickens, with seroconversion. Emergence of these new reassortant H9N2 avian influenza viruses indicates that these viruses can infect both chicken and goose and can produce different types of lesions in each species.
Keyword
MeSH Terms
Figure
Reference
-
1. Causey D, Edwards SV. Ecology of avian influenza virus in birds. J Infect Dis. 2008; 197:Suppl 1. S29–S33.
Article2. Cui L, Liu D, Shi W, Pan J, Qi X, Li X, Guo X, Zhou M, Li W, Li J, Haywood J, Xiao H, Yu X, Pu X, Wu Y, Yu H, Zhao K, Zhu Y, Wu B, Jin T, Shi Z, Tang F, Zhu F, Sun Q, Wu L, Yang R, Yan J, Lei F, Zhu B, Liu W, Ma J, Wang H, Gao GF. Dynamic reassortments and genetic heterogeneity of the human-infecting influenza A (H7N9) virus. Nat Commun. 2014; 5:3142.
Article3. Dong G, Luo J, Zhang H, Wang C, Duan M, Deliberto TJ, Nolte DL, Ji G, He H. Phylogenetic diversity and genotypical complexity of H9N2 influenza A viruses revealed by genomic sequence analysis. PLoS One. 2011; 6:e17212.
Article4. Edwards S. OIE laboratory standards for avian influenza. Dev Biol (Basel). 2006; 124:159–162.5. Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Li X, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Zhang Y, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013; 368:1888–1897.
Article6. Gibbs SE. Avian biology, the human influence on global avian influenza transmission, and performing surveillance in wild birds. Anim Health Res Rev. 2010; 11:35–41.
Article7. Guan Y, Peiris JS, Lipatov AS, Ellis TM, Dyrting KC, Krauss S, Zhang LJ, Webster RG, Shortridge KF. Emergence of multiple genotypes of H5N1 avian influenza viruses in Hong Kong SAR. Proc Natl Acad Sci U S A. 2002; 99:8950–8955.
Article8. Guan Y, Shortridge KF, Krauss S, Chin PS, Dyrting KC, Ellis TM, Webster RG, Peiris M. H9N2 influenza viruses possessing H5N1-like internal genomes continue to circulate in poultry in southeastern China. J Virol. 2000; 74:9372–9380.
Article9. Guo Y, Li J, Cheng X. [Discovery of men infected by avian influenza A (H9N2) virus]. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 1999; 13:105–108. Chinese.10. Guo YJ, Krauss S, Senne DA, Mo IP, Lo KS, Xiong XP, Norwood M, Shortridge KF, Webster RG, Guan Y. Characterization of the pathogenicity of members of the newly established H9N2 influenza virus lineages in Asia. Virology. 2000; 267:279–288.
Article11. Hatta M, Gao P, Halfmann P, Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science. 2001; 293:1840–1842.
Article12. Hatta M, Hatta Y, Kim JH, Watanabe S, Shinya K, Nguyen T, Lien PS, Le QM, Kawaoka Y. Growth of H5N1 influenza A viruses in the upper respiratory tracts of mice. PLoS Pathog. 2007; 3:1374–1379.
Article13. Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol. 2001; 146:2275–2289.
Article14. Huang R, Wang AR, Liu ZH, Liang W, Li XX, Tang YJ, Miao ZM, Chai TJ. Seroprevalence of avian influenza H9N2 among poultry workers in Shandong Province, China. Eur J Clin Microbiol Infect Dis. 2013; 32:1347–1351.
Article15. Krauss S, Webster RG. Avian influenza virus surveillance and wild birds: past and present. Avian Dis. 2010; 54:1 Suppl. 394–398.
Article16. Liu D, Shi W, Gao GF. Poultry carrying H9N2 act as incubators for novel human avian influenza viruses. Lancet. 2014; 383:869.
Article17. Liu H, Liu X, Cheng J, Peng D, Jia L, Huang Y. Phylogenetic analysis of the hemagglutinin genes of twenty-six avian influenza viruses of subtype H9N2 isolated from chickens in China during 1996-2001. Avian Dis. 2003; 47:116–127.
Article18. Mase M, Tanimura N, Imada T, Okamatsu M, Tsukamoto K, Yamaguchi S. Recent H5N1 avian influenza A virus increases rapidly in virulence to mice after a single passage in mice. J Gen Virol. 2006; 87:3655–3659.
Article19. Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, Castrucci MR, Donatelli I, Kawaoka Y. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol. 2000; 74:8502–8512.
Article20. Munster VJ, Fouchier RA. Avian influenza virus: of virus and bird ecology. Vaccine. 2009; 27:6340–6344.
Article21. Obenauer JC, Denson J, Mehta PK, Su X, Mukatira S, Finkelstein DB, Xu X, Wang J, Ma J, Fan Y, Rakestraw KM, Webster RG, Hoffmann E, Krauss S, Zheng J, Zhang Z, Naeve CW. Large-scale sequence analysis of avian influenza isolates. Science. 2006; 311:1576–1580.
Article22. Peiris M, Yuen KY, Leung CW, Chan KH, Ip PL, Lai RW, Orr WK, Shortridge KF. Human infection with influenza H9N2. Lancet. 1999; 354:916–917.
Article23. Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. Am J Epidemiol. 1938; 27:493–497.24. Shanmuganatham KK, Jones JC, Marathe BM, Feeroz MM, Jones-Engel L, Walker D, Turner J, Rabiul Alam SM, Kamrul Hasan M, Akhtar S, Seiler P, McKenzie P, Krauss S, Webby RJ, Webster RG. The replication of Bangladeshi H9N2 avian influenza viruses carrying genes from H7N3 in mammals. Emerg Microbes Infect. 2016; 5:e35.
Article25. Shi W, Li W, Li X, Haywood J, Ma J, Gao GF, Liu D. Phylogenetics of varied subtypes of avian influenza viruses in China: potential threat to humans. Protein Cell. 2014; 5:253–257.
Article26. Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. Avian flu: influenza virus receptors in the human airway. Nature. 2006; 440:435–436.27. Sun X, Xu X, Liu Q, Liang D, Li C, He Q, Jiang J, Cui Y, Li J, Zheng L, Guo J, Xiong Y, Yan J. Evidence of avian-like H9N2 influenza A virus among dogs in Guangxi, China. Infect Genet Evol. 2013; 20:471–475.
Article28. Sun Y, Pu J, Jiang Z, Guan T, Xia Y, Xu Q, Liu L, Ma B, Tian F, Brown EG, Liu J. Genotypic evolution and antigenic drift of H9N2 influenza viruses in China from 1994 to 2008. Vet Microbiol. 2010; 146:215–225.
Article29. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013; 30:2725–2729.
Article30. Uyeki TM, Chong YH, Katz JM, Lim W, Ho YY, Wang SS, Tsang TH, Au WW, Chan SC, Rowe T, Hu-Primmer J, Bell JC, Thompson WW, Bridges CB, Cox NJ, Mak KH, Fukuda K. Lack of evidence for human-to-human transmission of avian influenza A (H9N2) viruses in Hong Kong, China 1999. Emerg Infect Dis. 2002; 8:154–159.
Article31. Vandegrift KJ, Sokolow SH, Daszak P, Kilpatrick AM. Ecology of avian influenza viruses in a changing world. Ann N Y Acad Sci. 2010; 1195:113–128.
Article32. Vines A, Wells K, Matrosovich M, Castrucci MR, Ito T, Kawaoka Y. The role of influenza A virus hemagglutinin residues 226 and 228 in receptor specificity and host range restriction. J Virol. 1998; 72:7626–7631.
Article33. Wan H, Sorrell EM, Song H, Hossain MJ, Ramirez-Nieto G, Monne I, Stevens J, Cattoli G, Capua I, Chen LM, Donis RO, Busch J, Paulson JC, Brockwell C, Webby R, Blanco J, Al-Natour MQ, Perez DR. Replication and transmission of H9N2 influenza viruses in ferrets: evaluation of pandemic potential. PLoS One. 2008; 3:e2923.
Article34. Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992; 56:152–179.
Article35. Wu A, Su C, Wang D, Peng Y, Liu M, Hua S, Li T, Gao GF, Tang H, Chen J, Liu X, Shu Y, Peng D, Jiang T. Sequential reassortments underlie diverse influenza H7N9 genotypes in China. Cell Host Microbe. 2013; 14:446–452.
Article36. Xu C, Fan W, Wei R, Zhao H. Isolation and identification of swine influenza recombinant A/Swine/Shandong/1/2003 (H9N2) virus. Microbes Infect. 2004; 6:919–925.
Article37. Yu Q, Liu L, Pu J, Zhao J, Sun Y, Shen G, Wei H, Zhu J, Zheng R, Xiong D, Liu X, Liu J. Risk perceptions for avian influenza virus infection among poultry workers, China. Emerg Infect Dis. 2013; 19:313–316.
Article38. Zhang P, Tang Y, Liu X, Peng D, Liu W, Liu H, Lu S, Liu X. Characterization of H9N2 influenza viruses isolated from vaccinated flocks in an integrated broiler chicken operation in eastern China during a 5 year period (1998-2002). J Gen Virol. 2008; 89:3102–3112.
Article39. Zhou J, Wu J, Zeng X, Huang G, Zou L, Song Y, Gopinath D, Zhang X, Kang M, Lin J, Cowling BJ, Lindsley WG, Ke C, Peiris JS, Yen HL. Isolation of H5N6, H7N9 and H9N2 avian influenza A viruses from air sampled at live poultry markets in China, 2014 and 2015. Euro Surveill. 2016; 21:30331.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- H5 and H9 subtypes of Avian Influenza Viruses are Real Threat To Humans
- Genetic characterization of H9N2 avian influenza virus previously unrecognized in Korea
- Surveillance of wild birds for avian influenza virus in Korea
- Isolation and phylogenetic analysis of hemagglutinin gene of H9N2 influenza viruses from chickens in South China from 2012 to 2013
- Current situation and control strategies of H9N2 avian influenza in South Korea