J Periodontal Implant Sci.
2016 Feb;46(1):35-45. 10.5051/jpis.2016.46.1.35.
Prevalence of Porphyromonas gingivalis fimA genotypes in the peri-implant sulcus of Koreans assessed using a new primer
- Affiliations
-
- 1Department of Periodontology, School of Dentistry, Kyung Hee University, Seoul, Korea. yherr@khu.ac.kr
- 2Department of Periodontology, Institute of Oral Biology, School of Dentistry, Kyung Hee University, Seoul, Korea.
- 3Department of Maxillofacial Biomedical Engineering, School of Dentistry, Kyung Hee University, Seoul, Korea.
- KMID: 2391728
- DOI: http://doi.org/10.5051/jpis.2016.46.1.35
Abstract
- PURPOSE
Porphyromonas gingivalis fimA is a virulence factor associated with periodontal diseases, but its role in the pathogenesis of peri-implantitis remains unclear. We aimed to evaluate the relationship between the condition of peri-implant tissue and the distribution of P. gingivalis fimA genotypes in Koreans using a new primer.
METHODS
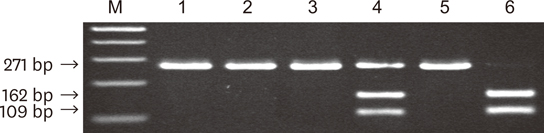
A total of 248 plaque samples were taken from the peri-implant sulci of 184 subjects. The control group consisted of sound implants with a peri-implant probing depth (PD) of 5 mm or less with no bleeding on probing (BOP). Test group I consisted of implants with a peri-implant PD of 5 mm or less and BOP, and test group II consisted of implants with a peri-implant PD of more than 5 mm and BOP. DNA was extracted from each sample and analyzed a using a polymerase chain reaction (PCR) with P. gingivalis-specific primers, followed by an additional PCR assay to differentiate the fimA genotypes in P. gingivalis- positive subjects.
RESULTS
The Prevalence of P. gingivalis in each group did not significantly differ (P>0.05). The most predominant fimA genotype in all groups was type II. The prevalence of type Ib fimA was significantly greater in test group II than in the control group (P<0.05).
CONCLUSIONS
The fimA type Ib genotype of P. gingivalis was found to play a critical role in the destruction of peri-implant tissue, suggesting that it may be a distinct risk factor for peri-implantitis.
MeSH Terms
Figure
Reference
-
1. Mombelli A, Lang NP. The diagnosis and treatment of peri-implantitis. Periodontol 2000. 1998; 17:63–76.
Article2. Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, et al. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001; 183:3770–3783.
Article3. Heydenrijk K, Meijer HJ, van der Reijden WA, Raghoebar GM, Vissink A, Stegenga B. Microbiota around root-form endosseous implants: a review of the literature. Int J Oral Maxillofac Implants. 2002; 17:829–838.4. de Oliveira GR, Pozzer L, Cavalieri-Pereira L, de Moraes PH, Olate S, de Albergaría Barbosa JR. Bacterial adhesion and colonization differences between zirconia and titanium implant abutments: an in vivo human study. J Periodontal Implant Sci. 2012; 42:217–223.
Article5. Mombelli A, Marxer M, Gaberthüel T, Grunder U, Lang NP. The microbiota of osseointegrated implants in patients with a history of periodontal disease. J Clin Periodontol. 1995; 22:124–130.
Article6. Amano A, Nakagawa I, Kataoka K, Morisaki I, Hamada S. Distribution of Porphyromonas gingivalis strains with fimA genotypes in periodontitis patients. J Clin Microbiol. 1999; 37:1426–1430.
Article7. Salcetti JM, Moriarty JD, Cooper LF, Smith FW, Collins JG, Socransky SS, et al. The clinical, microbial, and host response characteristics of the failing implant. Int J Oral Maxillofac Implants. 1997; 12:32–42.8. Rutar A, Lang NP, Buser D, Bürgin W, Mombelli A. Retrospective assessment of clinical and microbiological factors affecting periimplant tissue conditions. Clin Oral Implants Res. 2001; 12:189–195.
Article9. Botero JE, González AM, Mercado RA, Olave G, Contreras A. Subgingival microbiota in peri-implant mucosa lesions and adjacent teeth in partially edentulous patients. J Periodontol. 2005; 76:1490–1495.
Article10. Amano A. Molecular interaction of Porphyromonas gingivalis with host cells: implication for the microbial pathogenesis of periodontal disease. J Periodontol. 2003; 74:90–96.
Article11. Imamura T. The role of gingipains in the pathogenesis of periodontal disease. J Periodontol. 2003; 74:111–118.
Article12. Amano A, Sojar HT, Lee JY, Sharma A, Levine MJ, Genco RJ. Salivary receptors for recombinant fimbrillin of Porphyromonas gingivalis. Infect Immun. 1994; 62:3372–3380.
Article13. Nakagawa I, Amano A, Kuboniwa M, Nakamura T, Kawabata S, Hamada S. Functional differences among FimA variants of Porphyromonas gingivalis and their effects on adhesion to and invasion of human epithelial cells. Infect Immun. 2002; 70:277–285.
Article14. Enersen M, Nakano K, Amano A. Porphyromonas gingivalis fimbriae. J Oral Microbiol. 2013; 5:5.
Article15. Hamada N, Watanabe K, Arai M, Hiramine H, Umemoto T. Cytokine production induced by a 67-kDa fimbrial protein from Porphyromonas gingivalis. Oral Microbiol Immunol. 2002; 17:197–200.
Article16. Lee JY, Sojar HT, Bedi GS, Genco RJ. Porphyromonas (Bacteroides) gingivalis fimbrillin: size, amino-terminal sequence, and antigenic heterogeneity. Infect Immun. 1991; 59:383–389.
Article17. Nakagawa I, Amano A, Ohara-Nemoto Y, Endoh N, Morisaki I, Kimura S, et al. Identification of a new variant of fimA gene of Porphyromonas gingivalis and its distribution in adults and disabled populations with periodontitis. J Periodontal Res. 2002; 37:425–432.
Article18. Umeda JE, Missailidis C, Longo PL, Anzai D, Wikström M, Mayer MP. Adhesion and invasion to epithelial cells by fimA genotypes of Porphyromonas gingivalis. Oral Microbiol Immunol. 2006; 21:415–419.
Article19. Amano A, Kuboniwa M, Nakagawa I, Akiyama S, Morisaki I, Hamada S. Prevalence of specific genotypes of Porphyromonas gingivalis fimA and periodontal health status. J Dent Res. 2000; 79:1664–1668.
Article20. Nagano K, Abiko Y, Yoshida Y, Yoshimura F. Genetic and antigenic analyses of Porphyromonas gingivalis FimA fimbriae. Mol Oral Microbiol. 2013; 28:392–403.
Article21. Shin SI, Kwon YH, Park JB, Herr Y, Chung JH. Prevalence of fimA Genotypes of Porphyromonas gingivalis Strains in peri-implantitis patients. J Korean Acad Periodontol. 2005; 35:31–41.
Article22. Seo DK, Kwon YH, Park JB, Herr Y, Chung JH. Prevalence of fimA Genotypes of Porphyromonas gingivalis Strains in peri-implant sulcus. J Korean Acad Periodontol. 2005; 35:907–919.
Article23. Enersen M, Olsen I, Kvalheim Ø, Caugant DA. fimA genotypes and multilocus sequence types of Porphyromonas gingivalis from patients with periodontitis. J Clin Microbiol. 2008; 46:31–42.
Article24. Moon JH, Shin SI, Chung JH, Lee SW, Amano A, Lee JY. Development and evaluation of new primers for PCR-based identification of type II fimA of Porphyromonas gingivalis. FEMS Immunol Med Microbiol. 2011; 64:425–428.
Article25. Cohen J. A power primer. Psychol Bull. 1992; 112:155–159.
Article26. Moon JH, Herr Y, Lee HW, Shin SI, Kim C, Amano A, et al. Genotype analysis of Porphyromonas gingivalis fimA in Korean adults using new primers. J Med Microbiol. 2013; 62:1290–1294.
Article27. Zhao L, Wu YF, Meng S, Yang H, OuYang YL, Zhou XD. Prevalence of fimA genotypes of Porphyromonas gingivalis and periodontal health status in Chinese adults. J Periodontal Res. 2007; 42:511–517.
Article28. Missailidis CG, Umeda JE, Ota-Tsuzuki C, Anzai D, Mayer MP. Distribution of fimA genotypes of Porphyromonas gingivalis in subjects with various periodontal conditions. Oral Microbiol Immunol. 2004; 19:224–229.
Article29. Puig-Silla M, Dasí-Fernández F, Montiel-Company JM, Almerich-Silla JM. Prevalence of fimA genotypes of Porphyromonas gingivalis and other periodontal bacteria in a Spanish population with chronic periodontitis. Med Oral Patol Oral Cir Bucal. 2012; 17:e1047–53.
Article30. Nakano K, Kuboniwa M, Nakagawa I, Yamamura T, Nomura R, Okahashi N, et al. Comparison of inflammatory changes caused by Porphyromonas gingivalis with distinct fimA genotypes in a mouse abscess model. Oral Microbiol Immunol. 2004; 19:205–209.
Article31. Subramani K, Jung RE, Molenberg A, Hammerle CH. Biofilm on dental implants: a review of the literature. Int J Oral Maxillofac Implants. 2009; 24:616–626.32. Koyanagi T, Sakamoto M, Takeuchi Y, Ohkuma M, Izumi Y. Analysis of microbiota associated with peri-implantitis using 16S rRNA gene clone library. J Oral Microbiol. 2010; 2:2.
Article33. Laine ML, van Winkelhoff AJ. Virulence of six capsular serotypes of Porphyromonas gingivalis in a mouse model. Oral Microbiol Immunol. 1998; 13:322–325.
Article34. Rylev M, Kilian M. Prevalence and distribution of principal periodontal pathogens worldwide. J Clin Periodontol. 2008; 35:Suppl. 346–361.
Article35. van der Ploeg JR, Giertsen E, Lüdin B, Mörgeli C, Zinkernagel AS, Gmür R. Quantitative detection of Porphyromonas gingivalis fimA genotypes in dental plaque. FEMS Microbiol Lett. 2004; 232:31–37.36. Beikler T, Peters U, Prajaneh S, Prior K, Ehmke B, Flemmig TF. Prevalence of Porphyromonas gingivalis fimA genotypes in Caucasians. Eur J Oral Sci. 2003; 111:390–394.
Article37. Hayashi F, Okada M, Oda Y, Kojima T, Kozai K. Prevalence of Porphyromonas gingivalis fimA genotypes in Japanese children. J Oral Sci. 2012; 54:77–83.
Article38. Perez-Chaparro PJ, Rouillon A, Minet J, Lafaurie GI, Bonnaure-Mallet M. fimA genotypes and PFGE profile patterns in Porphyromonas gingivalis isolates from subjects with periodontitis. Oral Microbiol Immunol. 2009; 24:423–426.
Article39. Tamura K, Nakano K, Nomura R, Miyake S, Nakagawa I, Amano A, et al. Distribution of Porphyromonas gingivalis fimA genotypes in Japanese children and adolescents. J Periodontol. 2005; 76:674–679.
Article40. Nakagawa I, Amano A, Kimura RK, Nakamura T, Kawabata S, Hamada S. Distribution and molecular characterization of Porphyromonas gingivalis carrying a new type of fimA gene. J Clin Microbiol. 2000; 38:1909–1914.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Prevalence of fimA Genotypes of Porphyromonas gingivalis Strains in peri-implant sulcus
- Prevalence of fimA Genotypes of Porphyromonas gingivalis Strains in peri-implantitis patients
- Difference in Prevalence of fimA Genotypes of Porphyromonas gingivalis between Smoker and non-smoker Patients with Periodontitis
- Environmental factors regulating the expression of Porphyromonas gingivalis heat shock protein
- The relation between metabolic acid and the virulence of porphyromonas gingivalis serotype b