Transl Clin Pharmacol.
2017 Jun;25(2):74-84. 10.12793/tcp.2017.25.2.74.
Parameter estimation for sigmoid E(max) models in exposure-response relationship
- Affiliations
-
- 1Department of Clinical Pharmacology, Pusan National University Hospital, Busan 49241, Republic of Korea. dhlee97@gmail.com
- 2(Bio)Medical Research Institute, Pusan National University Hospital, Busan 49241, Republic of Korea.
- KMID: 2386764
- DOI: http://doi.org/10.12793/tcp.2017.25.2.74
Abstract
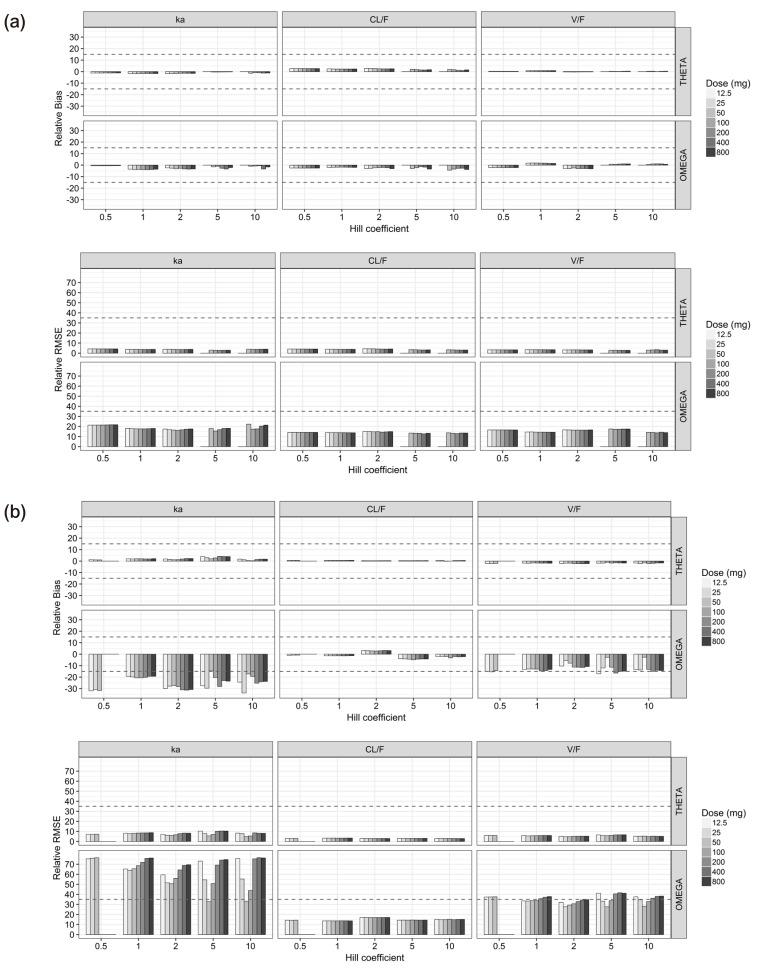
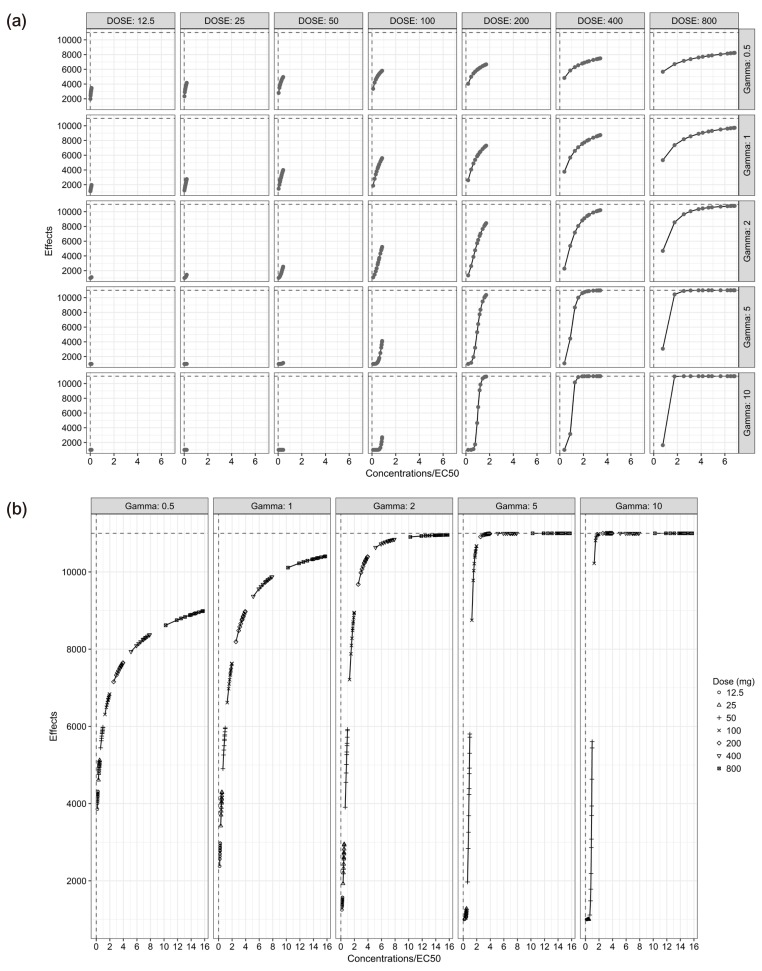
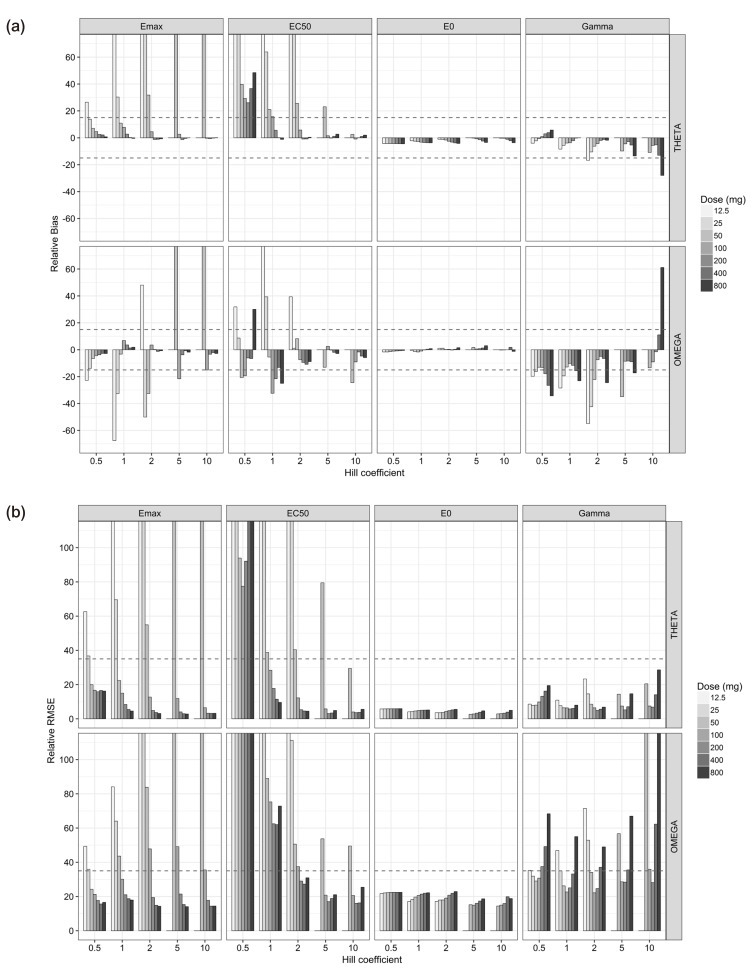
- The purpose of this simulation study is to explore the limitation of the population PK/PD analysis using data from a clinical study and to help to construct an appropriate PK/PD design that enable precise and unbiased estimation of both fixed and random PD parameters in PK/PD analysis under different doses and Hill coefficients. Seven escalating doses of virtual drugs with equal potency and efficacy but with five different Hill coefficients were used in simulations of single and multiple dose scenarios with dense sampling design. A total of 70 scenarios with 100 subjects were simulated and estimated 100 times applying 1-compartment PK model and sigmoid E(max) model. The bias and precision of the parameter estimates in each scenario were assessed using relative bias and relative root mean square error. For the single dose scenarios, most PD parameters of sigmoid E(max) model were accurately and precisely estimated when the C(max) was more than 85% of ECâ‚…â‚€, except for typical value and inter-individual variability of ECâ‚…â‚€ which were poorly estimated at low Hill coefficients. For the multiple dose studies, the parameter estimation performance was not good. This simulation study demonstrated the effect of the relative range of sampled concentrations to ECâ‚…â‚€ and sigmoidicity on the parameter estimation performance using dense sampling design.
Keyword
Figure
Reference
-
1. Mould DR, Upton RN. Basic concepts in population modeling, simulation, and model-based drug development. CPT Pharmacometrics Syst Pharmacol. 2012; 1:e6. DOI: 10.1038/psp.2012.4. PMID: 23835886.
Article2. Lee JY, Garnett CE, Gobburu JV, Bhattaram VA, Brar S, Earp JC, et al. Impact of pharmacometric analyses on new drug approval and labelling decisions: a review of 198 submissions between 2000 and 2008. Clin Pharmacokinet. 2011; 50:627–635. DOI: 10.2165/11593210-000000000-00000. PMID: 21895036.3. Wang Y, Bhattaram AV, Jadhav PR, Lesko LJ, Madabushi R, Powell JR, et al. Leveraging prior quantitative knowledge to guide drug development decisions and regulatory science recommendations: impact of FDA pharmacometrics during 2004-2006. J Clin Pharmacol. 2008; 48:146–156. DOI: 10.1177/0091270007311111. PMID: 18199891.
Article4. Lesko LJ, Schmidt S. Individualization of drug therapy: history, present state, and opportunities for the future. Clin Pharmacol Ther. 2012; 92:458–466. DOI: 10.1038/clpt.2012.113. PMID: 22948891.
Article5. Trivedi A, Lee RE, Meibohm B. Applications of pharmacometrics in the clinical development and pharmacotherapy of anti-infectives. Expert Rev Clin Pharmacol. 2013; 6:159–170. DOI: 10.1586/ecp.13.6. PMID: 23473593.
Article6. Holford NH, Sheiner LB. Understanding the dose-effect relationship: clinical application of pharmacokinetic-pharmacodynamic models. Clin Pharmacokinet. 1981; 6:429–453. PMID: 7032803.7. Dayneka NL, Garg V, Jusko WJ. Comparison of four basic models of indirect pharmacodynamic responses. J Pharmacokinet Biopharm. 1993; 21:457–478. PMID: 8133465.
Article8. Mould DR, Denman NG, Duffull S. Using disease progression models as a tool to detect drug effect. Clin Pharmacol Ther. 2007; 82:81–86. PMID: 17507925.
Article9. Goutelle S, Maurin M, Rougier F, Barbaut X, Bourguignon L, Ducher M, et al. The Hill equation: a review of its capabilities in pharmacological modelling. Fundam Clin Pharmacol. 2008; 22:633–648. DOI: 10.1111/j.1472-8206.2008.00633.x. PMID: 19049668.
Article10. Girgis S, Pai SM, Girgis IG, Batra VK. Pharmacodynamic parameter estimation: population size versus number of samples. AAPS J. 2005; 7:46. PMID: 16353905.
Article11. Pai SM, Girgis S, Batra VK, Girgis IG. Population pharmacodynamic parameter estimation from sparse sampling: effect of sigmoidicity on parameter estimates. AAPS J. 2009; 11:535–540. DOI: 10.1208/s12248-009-9131-2. PMID: 19629711.
Article12. Sun H, Ette EI, Ludden TM. On the recording of sample times and parameter estimation from repeated measures pharmacokinetic data. J Pharmacokinet Biopharm. 1996; 24:637–650. PMID: 9300354.
Article13. Ette EI, Chu HM, Godfrey CJ. Data supplementation: a pharmacokinetic/ pharmacodynamic knowledge creation approach for characterizing an unexplored region of the response surface. Pharm Res. 2005; 22:523–531. PMID: 15846459.14. Dutta S, Matsumoto Y, Ebling WF. Is it possible to estimate the parameters of the sigmoid Emax model with truncated data typical of clinical studies? J Pharm Sci. 1996; 85:232–239. PMID: 8683454.
Article15. Dragalin V, Hsuan F, Padmanabhan SK. Adaptive designs for dose-finding studies based on sigmoid Emax model. J Biopharm Stat. 2007; 17:1051–1070. PMID: 18027216.16. Wang TH, Yang M. Adaptive optimal designs for dose-finding studies based on sigmoid E-max models. J Stat Plan Inference. 2014; 144:188–197.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Drug interaction: focusing on response surface models
- Population pharmacodynamics of cilostazol in healthy Korean subjects
- The irtQ R package: a user-friendly tool for item response theory-based test data analysis and calibration
- Unobtrusive Estimation of Cardiorespiratory Fitness with Daily Activity in Healthy Young Men
- Evaluation through the Use of DSP2 Program for Sex Estimation by Measuring Human Hip Bones in a Joseon Dynasty Bone Collection